Identification and assessment of ferroptosis-related genes and their implication as therapeutic agents for pancreatic ductal adenocarcinoma (VSports)
Highlight box
Key findings
• Six ferroptosis-related genes (FRGs) (CYGB, EGR1, NTS, ATP6V1G2, SLC2A3, and SLC2A6) were identified as potential biomarkers and therapeutic targets in pancreatic ductal adenocarcinoma (PDAC).
• These genes are involved in oxidative stress, membrane transport, and metabolic regulation, and show significant prognostic value and druggability.
What is known and what is new?
• PDAC is a highly lethal cancer with limited diagnostic and therapeutic options V体育2025版. Ferroptosis plays a role in tumor suppression and drug sensitivity.
• This study systematically identifies and validates key FRGs with diagnostic, prognostic, and therapeutic relevance in PDAC, suggesting novel directions for ferroptosis-targeted treatments.
What is the implication, and what should change now?
• These findings support further research and clinical validation of FRGs as diagnostic markers and drug targets, paving the way for more effective, targeted PDAC therapies.
Introduction
Background
Pancreatic ductal adenocarcinoma (PDAC) is among the most aggressive forms of cancer and accounts for approximately 90% of all pancreatic malignancies. Global incidence and prevalence rate of PDAC is highly and rises and thus becomes a global health concern. The World Health Organization (WHO) and various cancer registries have documented a worrying trend: PDAC is now increasing not only in incidence but also in mortality and is predictable to be the 2nd most common cause of cancer-death in 2030, next only to the lung cancer (1). This dire amount puts into perspective the need for development of better diagnostic tools, management and treatment of PDAC VSports手机版.
Current projections indicate that new PDAC cases will be diagnosed in the United States only in 2024 with 60,100 and 49,770 deaths in the same year respectively. These statistics paint a rather bleak picture which depict the fact that the most of PDAC patients exist at late stages hence a situation that makes the disease more resistant to curative treatments (2,3). In low and middle-income countries (LMICs) there have also been increasing burden of PDAC, although the reported incidence value is lower than in high- and middle-income countries (HICs). Some of the causes for this disparity include concealment, ignorance, and restricted comer to diagnostic and health facilities (4). Treating PDAC remains a major clinical challenge primarily due to the difficulty in diagnosing the disease at an early stage. The pancreas is a concealed organ located in the abdominal cavity and in most cases, first symptoms of PDAC are not manifest. Symptoms such as jaundice, weight loss, and abdominal pain usually appear at advanced stages of the disease, when it has already progressed significantly and treatment options are limited (5). Surgery being the only curative therapeutic modality is possible in less than 20% of the patients due to vascular and metastatic involvement at presentation in the majority of patients (6) V体育安卓版.
PDAC remains a very significant problem in oncology; it is a problem that is experienced worldwide across all the demographic and geographic areas. The increasing rate and consistently unfavorable prognosis clearly indicate the inefficiency of available treatment methods and the need to develop new ones V体育ios版. This is due to the advancement in the knowledge pertaining to the molecular and cellular basis of PDAC where prospects for treatment are being revealed (7).
Ferroptosis (V体育平台登录)
Ferroptosis is a newly described type of programmed cell death that varied from the apoptosis, necrosis and autophagy. Ferroptosis is a distinct iron-dependent form of regulated cell death that differs from apoptosis, necrosis, and autophagy VSports最新版本. It is characterized by lipid peroxidation and associated membrane damage (8). Some of the identified regulators of ferroptosis are proteins that are involved in iron metabolism, GPX4, and the amino acid transporters of cystine-glutamate transporter SLC7A11 (system Xc−) (9).
Ferroptosis is initiated through iron-dependent redox reactions and Fenton chemistry, leading to lipid peroxidation. Glutathione depletion and inhibition of GPX4 are central to this process (9,10). System Xc which implies the transport of cysteine/glutamate is important in the importation of cysteine that is used in the synthesis of glutathione (11) V体育平台登录.
Ferroptosis-related genes (FRGs): they are the genes which are implicated in the process of ferroptosis which is a form of controlled cell death that actually varies from apoptosis and necrosis. Ferroptosis is one of the forms of cell death derived from iron-dependent lipid peroxidation and therefore, the oxidative stress related to it is toxic for the cells. This process has been reported to serve a number of physiological and pathological functions such as in cancer, neurodegenerative diseases, and tissue injury (12,13).
Ferroptosis as diagnostic and therapeutic agents
FRGs represent genes involved or implicated in regulating ferroptosis. These genes can either enhance or suppress the process and the expression levels can define how vulnerable the given cells are to ferroptotic cell death. In knowing the role of FRG’s is makes great value in research fields pertaining to cancer treat where the promotion of ferroptosis in cancer cells may be a cure (14). On one hand, it brings opportunity of being a therapeutic target through the selectively triggering of ferroptosis in cancer cells that have been known for their changed iron profile and increased level of oxidative stress (15). Targeting GPX4 in PDAC cells enhances ferroptosis by compromising their antioxidant defense, while inhibition of System Xc− reduces cystine import, leading to glutathione depletion and ferroptotic induction. This selective vulnerability could be utilized to kill the cancer cells more efficiently especially in those tumors which are now resistant to the traditional treatments. Ferroptosis, on the other hand, is more complicated and the induction of ferroptosis in normal cells or tissues may have critical side effects that are undesired, which makes the therapeutic approach more complicating (16).
Rationale and knowledge gap
Despite increasing interest in ferroptosis, its role in PDAC remains insufficiently characterized, particularly in terms of identifying specific FRGs that can serve as biomarkers or therapeutic targets. Previous studies have explored individual FRGs or general ferroptosis mechanisms in cancer, but few have applied a multi-step, integrative bioinformatics approach to systematically explore ferroptosis in PDAC using high-throughput datasets. While chemotherapy and radiotherapy are employed as adjuvant treatments, their impact on improving survival outcomes in PDAC has remained limited (17).
Furthermore, there is a lack of comprehensive analysis linking differentially expressed FRGs to patient prognosis, pathway involvement, and therapeutic relevance. Bridging this gap could yield actionable insights for ferroptosis-centered drug development and precision oncology in PDAC.
Objective
This study aimed to identify key FRGs with diagnostic, prognostic, and therapeutic relevance in PDAC using publicly available transcriptomic datasets. We employed a comprehensive computational pipeline encompassing differential expression analysis, functional enrichment [Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)], protein-protein interaction (PPI) network analysis, survival modeling, and drug-gene interaction profiling to prioritize candidate FRGs. Our goal was to highlight novel targets for ferroptosis modulation and provide a foundation for future experimental and clinical investigations in PDAC.
Methods
This work presents an approach to the prognostic and diagnostic markers of PDAC in relation to FRGs as depicted in Figure 1.
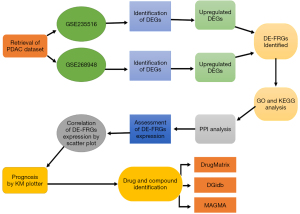
Gene Expression Omnibus (GEO) dataset and ferroptosis database (VSports在线直播)
The GEO (http://www.ncbi.nlm.nih.gov/geo/) is an global public source from microarray, next-generation sequencing, and other forms of high-throughput functional genomics data are archived and made freely available to the researchers worldwide from the contributions of the research community (18). It offers original data and rerun data and their descriptors in terms of different forms, formats, and strictly web-based tools and approaches that help users in searching the data, analyzing them, and presenting them visually. For this purpose, GEO is used to obtain the GSE235516 (19) and GSE268948 datasets involving the expression profile (20). The GSE235516 has six samples and is deposited in the Gene Expression Omnibus database, and Illumina NovaSeq 6000 (GPL24676) (21) is the sequencing platform used for this dataset, whereas the GSE268948 has also six samples have been downloaded from Gene Expression Omnibus database using Illumina NextSeq 500 (GPL18573) sequencing platform (22). As this study was conducted exclusively using publicly available transcriptomic datasets (GEO: GSE235516 and GSE268948) and did not involve any direct human or animal participants, ethical approval and informed consent were not required. Therefore, the ethical statement is not applicable in this context.
Identification of differentially expressed ferroptosis-related genes (DE-FRGs)
The DE for datasets GSE235516 and GSE268948 is in process using the DESeq2 package in R software (R Foundation for Statistical Computing, Vienna, Austria) (23). During this process, only gens with 2-fold change value >1 and adjusted P value <0.05 are grouped as DEGs and then isolated for further investigation. The process of analysis results to the construction of the Venn diagrams and heatmaps whereby the former is constructed using the ggplot2 package and the latter constructed using the ComplexHeatmap package (24,25). Furthermore, box plots are being generated using GraphPad Prism (v9. 3. 1, GraphPad Software Inc., La Jolla, CA, USA) to enhance data presentation, and for a better appreciation of the DE-FRGs under study (26,27).
GO and KEGG pathway enrichment analysis
The biological functions of the DE-FRGs are predicted using the GO enrichment analysis, in terms of the biological process (BP), cellular component (CC) and molecular function (MF) categories (28,29). KEGG analysis is used to locate these genes in the appropriate pathways, in order to create molecular interaction, reaction and relationship networks (30). For gene identification the package “org. Hs. eg. db” is used to decode gene symbol codes to Entrez ID (31). This analysis is performed using R software and for the assessment of enrichment, the subpackages of clusterProfiler and GOplot are used. The chosen level of significance is alpha equal to 0. 05 with the critical value of Q<0.2 (32). The data obtained from the plots are then plotted graphically with the help of the billion colors of the ‘ggplot2’ R package in other to get comprehensible graphical analysis of the plots (33). By these approaches, functional roles and pathway connectivity of DE-FRGs are examined global, and it would make easier to understand precisely the DE-FRGs’ roles in various BPs, cell components, and MFs (34,35). This approach allows addressing all the genes’ actions and their interactions in pairs and in general while understanding the system of molecular processes.
PPI analysis
PPI are defined as the assembly of two or more protein from different individuals held together by non-covalent bonds. In the area of protein micro array, PPI analysis is employed in the understanding of proteins functionality a connectivity, thus the potential role of the proteins in the BP. In this study, STRING version 11. 5 [https: The server-based software, STRING database (<www.string-db.org/>)] was used for the presentation and assessment of PPI networks (36,37). All differentially expressed FRGs are then imported into STRING to investigate the relationship of these genes. The analysis is performed using particular filter conditions, such as confidence score should be greater than or equal to 0.40, and, number of interactors should be less than 0, which makes the reported interaction more accurate (38,39). The subsequent results are then imported into Cytoscape (v 3. 9. 1) software for additional network analysis (40). In Cytoscape, the highly interconnected subnetworks of PPI are explored to find out the significant modules of PPIs (41). The molecular complex detection (MCODE) plug-in is used with default settings for screening and aims to find the densely connected sub-graphs that can be considered as functional protein complexes. Further, to list as well as rank the ten most central genes in the PPI network, the cytoHubba plug-in is applied to ascertain hub genes that might play significant roles in the BPs of interest (42,43).
V体育官网 - Assessment of DE-FRGs expression in cancers and normal tissues
To measure the expression of DE-FRGs in cancers and normal tissues, we used the DiffExp tool in the TIMER tool. Firstly, we listed target DE-FRGs based on literature research and FRG databases. The following genes were then investigated in great detail using TIMER, which provides various tools to analyze gene expression data in cancer (44,45). TIMER web portal was used and for the analysis the DiffExp module was applied in order to compare the levels of DE-FRGs in cancer tissue to the corresponding normal tissue. Through TIMER’s DiffExp module, users can analyze differential expression by choosing specific cancer types and key gene inputs (46). The module also includes determination of the level of significance, as well as comparison of expression levels using box-plots and other graphical displays. As for each DE-FRG, we have selected the most appropriate filter criteria which allowed us to define the reliability of the analysis. This was done through evaluation with TIMER to determine the p-values and fold changes of the samples to identify the number of genes that reported relative expression between cancer and normal samples. This strategy enables the collection of significant knowledge concerning the functions of DE-FRGs in cancer and tumorigenesis.
Correlation of DE-FRGs with scatter plot expression (VSports手机版)
Subsequently to enhance the reliability of the results, the expression values of DE-FRGs were cleaned and scaled using R software. To analyze the relation of DE-FRGs in different samples, scatter plot analysis (47). These plots were aimed at presenting what the analysis of DE-FRGs looked like in cancers and how it was in normal tissues, if there was any shift or pattern thereof was shown. Others statistical significant tests and other emerging trends were also utilized in establishing the extent and direction of the relationship depicted by the scatter plots, through the use of Pearson or Spearman Rank Order correlation coefficients.
KM plotter analysis of top prioritized DE-FRGs
The prognostic implication of DE-FRGs expression was evaluated by Kaplan-Meier Plotter (KM Plotter) database containing gene expression and survival information of PDAC patients (48). For assessment of overall survival (OS) and relapse-free survival (RFS), progression-free survival (PFS) and post-progression survival (PPS) the models were divided into high and low expression collections rendering to the median expression levels. KM plots were used to perform survival analysis while hazard ratios (HRs) with their corresponding 95% confidence intervals (CIs) and log-rank P values were estimated.
Drug and compound identification
The Drug signatures database integrated in the Enrichr resource (https://enrichr.webgestalt.org/; accessed on 6 September 2021) was used to identify drugs and compounds targeting the DE-FRGs, available from the following URL://maayanlab.cloud/Enrichr. The Drug and Compound Design within Enrichr identifies the relationships between gene sets and compounds, which is useful for drug discovery and drug repurposing (49).
Results
Identification of differentially expressed genes (DEGs)
Gene expression datasets GSE235516 and GSE268948 were analyzed to identify DEGs. GSE235516 have total 16,388 genes and identified DEGs are 3,131 as shown by volcano plot (Figure 2A). In total the up-regulated genes are 30 while down-regulated genes are 1,374. GSE268948 dataset have total 16,729 genes and the identified DEGs are 1,774 (Figure 2B), the upregulated genes in these DEGs are 76 and down-regulated genes are 944. Table S1 cataloged the up-regulated genes acquired from both the datasets.
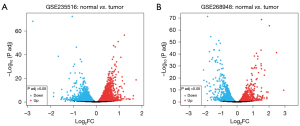
Identification of FRGs
A gene set of ferroptosis related genes from FerrDb database was obtained and compared with the DEGs of GSE235516. The result in identification of 5 upregulated FRGs and 52 downregulated FRGs (Figure 3A). These FRGs genes are listed in GSE268948 where we found 1 is upregulated and 57 is down-regulated DE-FRGs (Figure 3B). These FRGs are shown in a Venn diagram as shown in the Figure 3 below. Furthermore, we generated a heatmap to visualize the upregulated differential expression profiles of these six FRGs in the two datasets, as shown in Figure 3C.
Gene Ontology and KEGG pathways analysis of DE-FRGs
The research results GO and KEGG pathways and compares the DE-FRGs, and all the results are reorganized into several categories by BPs.
GO-BP
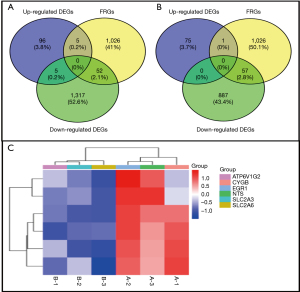
Based on the impact assessment, the results show several BPs that are largely affected by DE-FRGs. These processes incorporate the detailed molecular procedures and interactions that are involved in cellular functions and pathways such as Fructose transmembrane, Dehydroascorbic acid Transsport, Glucose transport and Vitamin metabolism which are closely linked to ferroptosis (Figures S1,S2A, Table 1).
Table 1
| Ontology | IDs | Description | Gene ratio | BgRatio | P value | P adjust | q value |
|---|---|---|---|---|---|---|---|
| BP | GO:0030198 | Extracellular matrix organization | 25/165 | 395/18,866 | 8.85E−15 | 1.27E−11 | 1.07E−11 |
| BP | GO:0043062 | Extracellular structure organization | 25/165 | 396/18,866 | 9.37E−15 | 1.27E−11 | 1.07E−11 |
| BP | GO:0050678 | Regulation of epithelial cell proliferation | 17/165 | 395/18,866 | 7.16E−08 | 6.44E−05 | 5.45E−05 |
| BP | GO:0050673 | Epithelial cell proliferation | 17/165 | 453/18,866 | 4.98E−07 | <0.001 | 0.000271 |
| BP | GO:0022617 | Extracellular matrix disassembly | 8/165 | 82/18,866 | 5.94E−07 | <0.001 | 0.000271 |
| BP | GO:0007369 | Gastrulation | 11/165 | 189/18,866 | 8.79E−07 | <0.001 | 0.000334 |
| BP | GO:0098773 | Skin epidermis development | 8/165 | 90/18,866 | 1.22E−06 | <0.001 | 0.000397 |
| BP | GO:0001704 | Formation of primary germ layer | 9/165 | 127/18,866 | 1.77E−06 | <0.001 | 0.000504 |
| BP | GO:0090287 | Regulation of cellular response to growth factor stimulus | 13/165 | 310/18,866 | 3.52E−06 | 0.001 | 0.000894 |
| BP | GO:0008544 | Epidermis development | 16/165 | 477/18,866 | 4.66E−06 | 0.001 | 0.001065 |
| CC | GO:0062023 | Collagen-containing extracellular matrix | 22/169 | 427/19,559 | 1.94E−11 | 4.41E−09 | 3.87E−09 |
| CC | GO:0005788 | Endoplasmic reticulum lumen | 15/169 | 308/19,559 | 7.73E−08 | 8.77E−06 | 7.69E−06 |
| CC | GO:0005604 | Basement membrane | 9/169 | 103/19,559 | 2.72E−07 | 2.05E−05 | 1.8E−05 |
| CC | GO:0005581 | Collagen trimer | 8/169 | 87/19,559 | 8.61E−07 | 4.89E−05 | 4.28E−05 |
| CC | GO:0005911 | Cell-cell junction | 15/169 | 493/19,559 | 2.58E−05 | 0.001 | 0.001025 |
| CC | GO:0098644 | Complex of collagen trimers | 3/169 | 21/19,559 | <0.001 | 0.03 | 0.024924 |
| CC | GO:0016324 | Apical plasma membrane | 10/169 | 361/19,559 | 0.001 | 0.04 | 0.034402 |
| CC | GO:0045177 | Apical part of cell | 11/169 | 433/19,559 | 0.001 | 0.04 | 0.034704 |
| CC | GO:0030027 | Lamellipodium | 7/169 | 201/19,559 | 0.002 | 0.046 | 0.039983 |
| CC | GO:0005796 | Golgi lumen | 5/169 | 103/19,559 | 0.002 | 0.046 | 0.039983 |
| MF | GO:0005201 | Extracellular matrix structural constituent | 13/165 | 169/18,352 | 4.26E−09 | 1.53E−06 | 1.38E−06 |
| MF | GO:0030020 | Extracellular matrix structural constituent conferring tensile strength | 7/165 | 41/18,352 | 7.27E−08 | 1.31E−05 | 1.17E−05 |
| MF | GO:0008201 | Heparin binding | 10/165 | 169/18,352 | 3.07E−06 | <0.001 | 0.000331 |
| MF | GO:0019838 | Growth factor binding | 8/165 | 136/18,352 | 3.2E−05 | 0.002 | 0.001972 |
| MF | GO:0048018 | Receptor ligand activity | 15/165 | 487/18,352 | 3.49E−05 | 0.002 | 0.001972 |
| MF | GO:0030546 | Signaling receptor activator activity | 15/165 | 492/18,352 | 3.92E−05 | 0.002 | 0.001972 |
| MF | GO:0005539 | Glycosaminoglycan binding | 10/165 | 232/18,352 | 4.86E−05 | 0.002 | 0.001972 |
| MF | GO:0017134 | Fibroblast growth factor binding | 4/165 | 23/18,352 | 4.88E−05 | 0.002 | 0.001972 |
| MF | GO:1901681 | Sulfur compound binding | 10/165 | 262/18,352 | <0.001 | 0.005 | 0.004781 |
| MF | GO:0005178 | Integrin binding | 7/165 | 144/18,352 | <0.001 | 0.01 | 0.010585 |
BgRatio: the number of genes inside each geneset. BP, biological process; CC, cellular component; DEGs, differentially expressed genes; GO, Gene Ontology; MF, molecular function.
GO-CC
Through our analysis, we determined that the following CCs are most impacted by the expression of DE-FRGs. Furthermore, this analysis provides a clear understanding of the cellular structures and organelles in which these genes are active. The results show that DE-FRGs are primarily enriched in the cell membrane, mitochondria, and endoplasmic reticulum organelles that are essential for the execution of ferroptosis (Figures S1,S2B, Table 1).
GO-MF
The MF of gene ontology of DE-FRGs involved in encoding proteins that are involved in biochemical functions including enzyme catalysis, ion binding and molecular transport. These functions are essential in controlling the onset and execution of ferroptosis, thus placing DE-FRGs at the center of a pro-cell survival/pro-cell death axis (Figures S1,S2C, Table 1).
KEGG pathways
This part of the analysis relates gene expression changes to other BPs to show the function of DE-FRGs in the large network of cellular signaling pathways. Of the 24 pathways, cancer, Rap1 signaling pathway, TGF-beta signaling pathway, neurodegenerative diseases and metabolic disorders are enriched, these indicates that genes involved in ferroptosis are implicated in a wide range of BPs (Figure S2D, Table 2). This analysis and categorization based on GO and KEGG pathways help to understand the functional consequences of DE-FRGs and also provide a basis for further studies regarding possible treatments and interventions in diseases associated with ferroptosis.
V体育官网入口 - Table 2
| ID | Description | GeneRatio | BgRatio | P value | P adjust | q value |
|---|---|---|---|---|---|---|
| hsa04974 | Protein digestion and absorption | 7/84 | 103/8,223 | 8.10083E−05 | 0.02 | 0.015604754 |
| hsa05219 | Bladder cancer | 4/84 | 41/8,223 | <0.001 | 0.07 | 0.062894149 |
| hsa05205 | Proteoglycans in cancer | 8/84 | 205/8,223 | 0.001 | 0.07 | 0.062894149 |
| hsa04015 | Rap1 signaling pathway | 8/84 | 210/8,223 | 0.001 | 0.07 | 0.062894149 |
| hsa04512 | ECM-receptor interaction | 5/84 | 88/8,223 | 0.002 | 0.08 | 0.076672911 |
| hsa04350 | TGF-beta signaling pathway | 5/84 | 96/8,223 | 0.003 | 0.10 | 0.091566738 |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 6/84 | 143/8,223 | 0.003 | 0.10 | 0.091566738 |
| hsa04722 | Neurotrophin signaling pathway | 5/84 | 119/8,223 | 0.007 | 0.19 | 0.174751601 |
| hsa04926 | Relaxin signaling pathway | 5/84 | 129/8,223 | 0.01 | 0.23 | 0.216322855 |
| hsa05323 | Rheumatoid arthritis | 4/84 | 93/8,223 | 0.01 | 0.26 | 0.23734025 |
| hsa00100 | Steroid biosynthesis | 2/84 | 20/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa01522 | Endocrine resistance | 4/84 | 98/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa05226 | Gastric cancer | 5/84 | 149/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 4/84 | 100/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa05165 | Human papillomavirus infection | 8/84 | 331/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa04625 | C-type lectin receptor signaling pathway | 4/84 | 104/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa04390 | Hippo signaling pathway | 5/84 | 157/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa04330 | Notch signaling pathway | 3/84 | 59/8,223 | 0.02 | 0.26 | 0.23734025 |
| hsa04060 | Cytokine-cytokine receptor interaction | 7/84 | 295/8,223 | 0.03 | 0.33 | 0.311162057 |
| hsa04924 | Renin secretion | 3/84 | 69/8,223 | 0.03 | 0.34 | 0.320271747 |
| hsa04744 | Phototransduction | 2/84 | 29/8,223 | 0.04 | 0.35 | 0.321309426 |
| hsa05206 | MicroRNAs in cancer | 7/84 | 310/8,223 | 0.04 | 0.36 | 0.338087313 |
| hsa04380 | Osteoclast differentiation | 4/84 | 128/8,223 | 0.04 | 0.37 | 0.348813924 |
| hsa04142 | Lysosome | 4/84 | 132/8,223 | 0.046 | 0.39 | 0.367349712 |
GeneRatio: the ratio of the number of genes in this GO word to the total number of genes in this category; BgRatio: the number of genes inside each geneset. DE-FRGs, differentially expressed ferroptosis-related genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
According to Gene Ontology: SLC2A3 is involved in the BP glucose import across plasma membrane. SLC2A3, and SLC2A6 belong to the BP hexose transmembrane transport (GO: 0008645). Early growth response 1 (EGR1) belongs to the BP cellular response to heparin SLC2A6 belongs to the BP fructose transmembrane transport (50) and SLC2A3 belong to the BP glucose transmembrane (51). From MGI Mammalian phenotype: several MP’s were noted in EGR1 KO mice: the phenotype abnormal circulating tumor necrosis factor level MP: 0008552. The phenotype abnormal liver physiology MP: EGR1 KO mice and in CYGB 0000609 was noticed. The phenotype abnormal circulating HDL cholesterol level MP: it is noteworthy that 0000184 was found to be increased in ATP6V1G2 KO mice. The phenotype increased incidence of tumors by chemical induction MP: the analysis showed that 0004499 was up regulated in EGR1 and CYGB KO mice. The phenotype abnormal physiological response to xenobiotic MP: the down-regulation of gene 0008872 was seen in EGR1 and NTS KO mice. From KEGG: ATP6V1G2 is one of the genes of the KEGG pathway entitled collecting duct acid secretion. ATP6V1G2 can also be attributed the gene product of which is involved in KEGG pathway-epithelial cell signaling in Helicobacter pylori infection. From the above analysis, the gene product EGR1 belongs to the KEGG pathway GnRH signaling pathway. ATP6V1G2 is implicated in the KEGG pathway that is associated with Vibrio cholerae infection. The gene product ATP6V1G2 is involved in Synaptic vesicle cycle as described in KEGG pathway map shown in Figure 4A,4B.
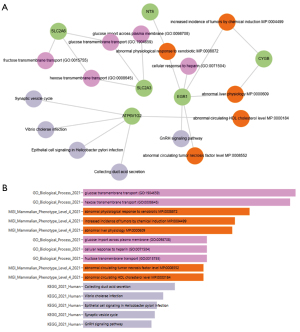
This figure highlights the involvement of target genes in metabolic transport, xenobiotic response, liver function, and signaling pathways, suggesting potential roles in disease and cellular regulation.
The KEGG pathway enrichment analysis revealed that the DE-FRGs are involved in several biologically significant pathways. Notably, genes such as ATP6V1G2, SLC2A3, and EGR1 showed enrichment in the TGF-β signaling pathway, Rap1 signaling pathway, ECM-receptor interaction, and neurotrophin signaling. These pathways are closely associated with tumor progression, cell proliferation, metabolic reprogramming, and modulation of the tumor microenvironment in PDAC. For example, SLC2A3 was linked to glucose transport and metabolic regulation, while EGR1 participated in cellular stress responses and inflammatory signaling. These pathway associations further support the hypothesis that FRGs have functional implications beyond cell death and may influence broader oncogenic networks in pancreatic cancer.
PPI network analysis and identification of hub genes
The six selected genes, which are CYGB, EGR1, NTS, ATP6V1G2, SLC2A3 and SLC2A6 were prioritized based on the integration of data from GeneMANIA and STRING, which are two different but related interaction databases. The prioritization was based on the strength of interactions, co-expression profiles and spatial relationships between the genes since all the interaction scores were obtained from both the databases. Initially, we looked for the overlap with other databases to find the genes that interact, co-expressed or physically associated in both the databases. To further ensure that the results were accurate, we only used gene pairs that were common in both GeneMANIA and STRING. For each of the common gene pairs, the interaction types and the scores were obtained from both the GeneMANIA and STRING databases (Figure 5A,5B). More emphasis was placed on gene pairs that have demonstrated significant interaction in both the databases. These scores were then added to give a total score for each gene pair for GeneMANIA and STRING. The interaction between DE-FRGs is illustrated in the Figure 5C.
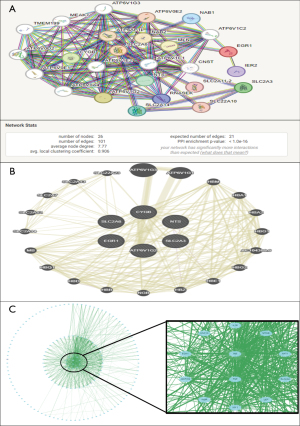
Assessment of DE-FRGs expression in cancers and normal tissues
In order to provide a systemic evaluation of the DE-FRGs’ expression patterns in various cancers, we used the DiffExp module of the TIMER database which is based on RNA-seq data from TCGA. We have identified six DE-FRGs, including CYGB, NTS, EGR1, ATP6V1G2, SLC2A3, and SLC2A6 because of their roles in ferroptosis and cancer biology as illustrated in Figures S3,S4. The comparison results showed the up- or down-regulation of the expression of the identified proteins in normal against tumor tissues in various cancers. Notably, CYGB and EGR1 were found to be upregulated in tumors and might be involved in tumorigenesis; while ATP6V1G2 and SLC2A6 were downregulated and may play a tumor-suppressive role via ferroptosis. Based on these observations, there is a need to further explore these DE-FRGs’ capability to be used as biomarkers in cancer diagnosis or prognosis. Since TIMER did not include PDAC data, the fact that there is a high degree of concordance in the expression of other cancers means that such mechanisms may be present in PDAC and other cancers not included in this analysis. This calls for more research on their mechanistic function and as the predictors of patients’ prognosis, treatment modalities, and therapeutic targets.
Correlation of DE-FRGs with scatter plot expression (VSports最新版本)
In the case of six DE-FRGs upregulated particularly, CYGB, EGR1, NTS, ATP6V1G2, SLC2A3, and SLC2A6, a scatter plot was used to analyze the relationship of expression levels. Pearson correlation coefficient which is known as R value, quantified the strength and direction of the linear relation between these genes. A higher R value meant that there was a positive association which means that if one gene expression is higher it is likely that the others will also have higher expression levels. It was also necessary to check the probability of these correlations by using the P value where P≤0. 05 was considered significant implying the correlation was real and not by chance as shown in Figure 6.
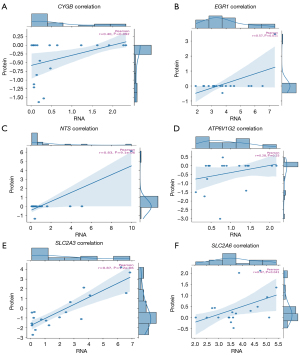
Each subplot shows a regression line with a 95% CI (shaded area), along with the Pearson correlation coefficient (r) and corresponding P value:
- CYGB: moderate positive correlation (r=0.40, P=0.092), not statistically significant.
- EGR1: significant positive correlation (r=0.57, P=0.01).
- NTS: strong and highly significant correlation (r=0.83, P=9.1e−06).
- ATP6V1G2: weak, non-significant correlation (r=0.28, P=0.25).
- SLC2A3: very strong and significant correlation (r=0.87, P=2e−06).
- SLC2A6: significant positive correlation (r=0.47, P=0.041).
These results indicate that most of the selected genes, particularly NTS, SLC2A3, and EGR1, exhibit a strong concordance between RNA and protein expression, supporting their potential as robust biomarkers or functional targets.
KM plotter analysis of top prioritized DE-FRGs
For assessing prognostic value of DE-FRGs in PDAC, and their correlation with CYGB, EGR1, NTS, ATP6V1G2, SLC2A3 & SLC2A6, KM plotter was employed and to ensure correct results, KM plots were generated using “all probe sets per gene” as shown in "VSports在线直播" Figure 7 and Table 3. In the context of a KM plot, the HR the P value checks the significance of the null hypothesis that there is no difference in the probability of survival between groups since a P value of ≤0.05 is considered significant. The false discovery rate (FDR) is a statistical measure to mitigate the probability of making at least one false positive while conducting multiple comparisons. It means that half of the population of a given cohort has died from the event, and it is also possible to present low and high median survival which means that the probability of typical survival for a certain biomarker or variable is different. As depicted in "VSports在线直播" Figure 7, these metrics offer a combined view of the survival differences between cohorts.
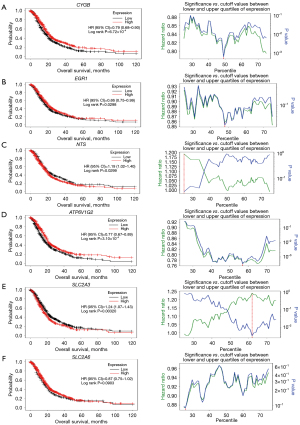
Table 3
| Gene symbol | Hazard ratio | P value | FDR | Median survival (low cohort) | Median survival (high cohort) |
|---|---|---|---|---|---|
| CYGB | 0.79 | <0.001 | 0.0281 | 16.77 | 20.13 |
| EGR1 | 0.86 | 0.03 | 0.199 | 17.07 | 19.9 |
| NTS | 1.19 | 0.03 | 0.851 | 19.9 | 18.1 |
| ATP6V1G2 | 0.77 | <0.001 | 0.00209 | 16.07 | 20.33 |
| SLC2A3 | 1.24 | 0.003 | 0.0426 | 20.0 | 16.23 |
| SLC2A6 | 0.87 | 0.09 | 0.522 | 17.03 | 19.27 |
Hazard ratio: a measure of the size of the disparity between the two curves in the Kaplan-Meier plot. DE-FRGs, differentially expressed ferroptosis-related genes; FDR, false discovery rate.
Drugs and compounds identification
The drugs and compounds targeting ferroptosis in the DE-FRGs were screened using the P value ranking strategy. In the analysis of DE-FRGs, three key categories of drugs were identified: Drug Matrix, Drug-Gene Interaction Database (DGIdb) targets, Multi-marker Analysis of Genomic Annotation (MAGMA) drugs and disease. Drug Matrix—a tool that can help to determine which compounds affect the expression of DE-FRGs, based on the interactions between drugs and gene expression profiles, i.e. shown in Figure 8A. DGIdb, a drug-target database, provides information on known and predicted interactions of these genes with drugs, indicating possible candidates for targeted therapy (Figure 8B). MAGMA drugs and diseases allow assessing the correlation between the analyzed genes and diseases as shown in Figure 8C.
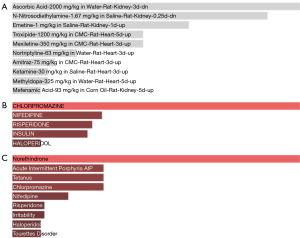
Drugs and chemicals that target ferroptosis in DE-FRGs were prioritized via a P value ranking. During the DE-FRG study, three main drug categories emerged as significant: Drug matrix, DGIdb targets, and MAGMA medications, and associated diseases with each medication. They are categorized as follows and each of them signifies a different strategy employed to screen chemicals that modulate ferroptosis-related processes.
DE-FRG expression is best understood from the Drug Matrix, an evaluation tool that analyzes chemical reactions with gene expression patterns. This way, the researchers may establish how these and other substances modify the probe fermoposis connected genes to understand the potential therapeutic methodologies. Figure 8 is on chemicals and affiliation to gene expression profiles.
The provided data from the DGIdb resource encompass both already established and potentially prove interaction between medicines and the considered genes. DGIdb helps to search for prospective drug candidates that could accurately interact with DE-FRGs and consequently aids in the development of targeted drugs. Figure 8B depicts the interactions using DGIdb.
MAGMA facilitates the investigation of the relationship between genes of interest and various disorders. This resource not only finds possible medications, but also shows the relationships between DE-FRGs and other disorders, give a comprehensive overview of the therapeutic landscape. The MAGMA analysis in Figure 8C illustrates the connection between these genes and illnesses, pointing to potential treatment approaches.
Through drug-gene interaction analysis using the Enrichr platform, several therapeutic compounds were identified that may target the prioritized DE-FRGs. Among the top candidates were vorinostat and trichostatin A, both histone deacetylase inhibitors known to induce ferroptosis and modulate tumor cell survival. Additionally, wortmannin and LY-294002, inhibitors of the PI3K/AKT signaling pathway, were linked to genes such as CYGB and SLC2A3, implicating their role in oxidative stress regulation and ferroptotic vulnerability. These compounds have been studied in preclinical cancer models and could offer a foundation for repurposing ferroptosis-targeted therapies in PDAC. The identification of these agents provides translational value and highlights potential avenues for therapeutic intervention based on ferroptosis modulation.
Discussion
PDAC is one of the most toxic malignant neoplasms mainly because of the high aggressiveness and a rather late stage of diagnosis (52). Due to the fact that PDAC develops silently and is only detected when the condition is already chronic, early diagnosis and treatment are hardly possible, and what is more important, therapeutic measures in case of PDAC are not very effective and prognosis is quite severe (53). Surgical resection, chemotherapy, and radiotherapy remain the mainstay of the current treatment which offers minimal survival benefit, thereby underlining the importance of discovery of novel therapeutic targets and better understanding of the disease biology of PDAC (54).
With the growing focus on the regulation of ferroptosis, the ferroptosis-inducing reagents (FINs) are becoming more and more important in tumor treatment.
To our knowledge, this study presents a novel integrative bioinformatics framework that combines differential gene expression analysis, functional enrichment (GO and KEGG), network mapping, correlation analyses, survival modeling, and drug signature mapping to identify and prioritize ferroptosis-related therapeutic targets in PDAC. While each of these components has individually been applied in previous studies, there is limited precedent for their combined use in a single, streamlined pipeline leading from gene identification to drug-target prediction. This layered approach enhances the robustness of biomarker prioritization by capturing both functional relevance and therapeutic tractability. The uniqueness of this methodology lies in its comprehensive nature, which bridges molecular discovery with translational drug insight, offering a reproducible framework for early-stage drug development in ferroptosis-centered cancer therapy. We believe this multi-step strategy can serve as a blueprint for future studies aimed at uncovering precision targets in similarly aggressive and treatment-resistant cancers.
In this study, a multifaceted bioinformatics analyses were pursued to define the list of ferroptosis-related biomarkers with the clinical relevance to PDAC prognosis. We found 76 upregulated DEGs from GSE235516 and GSE268948 datasets acquired from GEO resource, and the upregulated genes were processed as because of their high expression rate in cancers (55). The Pearson correlation analysis between prioritized DEGs and FRGs identified six probable genes CYGB, EGR1, NTS, ATP6V1G2, SLC2A3, and SLC2A6 involved in PDAC development.
GO and KEGG pathway analysis were conducted in order to understand the biological functions as well as the signaling pathways involved in DEGs. The GO analysis identified that these genes are involved in significant cellular processes such as the response to oxidative stress, iron homeostasis, and lipid metabolic process and, not surprisingly, since these pathways are all linked with the ferroptosis process. The MF enrichment analysis also gave similar results with some of the pathways such as the p53 signaling which has an active role in the regulation of ferroptosis. To reinforce these findings an integrated approach was taken with GeneMANIA and STRING databases to consider the possible role of six prominent DE-FRGs. This made it possible to view the fine network of the connectivity of genes as well as the co expression and physical relationship of these genes. From the analysis of the interactions of the genes profiled in the network, adequate evidence to assert that the genes are part of the large extended molecular regulatory network for ferroptosis in PDAC, were gathered. Among them, the cytoglobin (CYGB) had a very high degree centrality which may be attributed to the fact that it plays a role in oxidative stress and iron metabolism that is involved in ferroptosis. EGR1 which takes part in cell cycling and cell death process intimidate a relatively significant correlation and may be rated as one of the important additional and other candidates highlighted in the current investigation.
Functional enrichment analysis revealed that the identified FRGs are significantly involved in several critical molecular pathways. Notably, KEGG pathway analysis indicated enrichment in TGF-β signaling, Rap1 signaling, ECM-receptor interaction, neurotrophin signaling, and collecting duct acid secretion pathways. These pathways are known to influence cellular processes such as proliferation, apoptosis resistance, and immune modulation in pancreatic cancer. Furthermore, drug-gene interaction analysis using Enrichr identified several candidate compounds with potential therapeutic relevance. Among the top-ranked agents were vorinostat (a histone deacetylase inhibitor), wortmannin (a PI3K inhibitor), trichostatin A, and LY-294002, all of which have documented roles in modulating oxidative stress, cell cycle progression, or ferroptosis-related signaling. These findings suggest that the identified DE-FRGs not only participate in PDAC progression but may also serve as actionable targets for drug repurposing and the development of ferroptosis-focused therapies.
Particularly, according to the interference and function loss of these DE-FRGs, the TIMER database based on TCGA’s RNA-seq data was adopted to analyze the expression patterns of them in various cancers. These data suggest that these genes are up-regulated or down-regulated in several cancers thus, ferroptosis regulation is not cancer specific to PDAC. For this reason, the present genes might have possibilities in other types of cancers (56).
The Kaplan-Meier survival analysis was used with the purpose of evaluating the role of these DE-FRGs as prognostic factors in PDAC (57). According to the outcome of gene expression analysis, CYGB, EGR1, NTS, ATP6V1G2, SLC2A3, and SLC2A6 were found to be significantly associated with OS of PDAC patients. Most importantly, there were higher expressions of some genes that were related to poorer prognosis, thereby supporting their role as prognosis biomarkers (58). The findings of the current study is worthy as it establishes the possibility of exploiting ferroptosis signaling in PDAC for therapeutic advancement. Additionally, the study identified therapeutic agents that can affect these genes associated with ferroptosis. By analyzing drug-gene interactions, we categorized these agents into three primary groups: Drug Matrix, DGIdb drug targets and MAGMA associated drugs (59,60). The identification of these drugs has therefore presented a good platform for the development of therapeutic interventions especially when it comes to pharmacogenomics. From these studies, it may be postulated that modulating ferroptosis-associated pathways might improve the effectiveness of standard treatments or even define new therapeutic strategies in PDAC with regard to the molecular characteristics of the tumor (61,62). A related and important direction that needs further investigation is the interactions between ferroptosis and other forms of cell death, including apoptosis and necroptosis in PDAC. Studying these pathways may identify novel targets and rational poly-therapy that uses these pathways to induce cell death and overcome resistance leading to better prognosis for patients.
Furthermore, the fact that PDAC tumors are quite heterogeneous poses a major problem in applying these observations in the clinic. At the molecular level tumor heterogeneity implies that the level of DE-FRGs may also differ between patients and therefore influence the response to targeted therapies. All subsequent investigations should therefore be directed towards assessing the applicability of these genes in biomarker or diagnostic techniques that will help in differentiating patients, and thus offer better treatment regimens.
Our findings underscore a strong potential link between ferroptosis and PDAC. The identified DE-FRGs such as CYGB, EGR1, and SLC2A3 are associated with oxidative stress, iron regulation, and metabolic pathways known to influence ferroptotic activity. Given that PDAC cells often exhibit elevated reactive oxygen species and disrupted iron homeostasis, targeting ferroptosis presents a compelling therapeutic avenue. This study not only provides insight into the molecular underpinnings of ferroptosis in PDAC but also highlights promising biomarkers and druggable targets that warrant further experimental validation.
This study presents a comprehensive in silico investigation of FRGs in PDAC, integrating multiple bioinformatics tools to identify potential biomarkers and therapeutic targets. A key strength lies in the multi-step analysis approach, combining gene expression profiling, pathway enrichment, interaction networks, survival modeling, and drug-gene interaction prediction. However, the study is not without limitations. The analysis is based on publicly available transcriptomic data, and no experimental validation was performed to confirm the functional roles of the identified genes. The sample size in the GEO datasets is limited, and an independent PDAC-specific validation cohort was not included. Additionally, while we identified potential therapeutic agents, pharmacological testing was beyond the scope of this work. Future studies should include laboratory validation and clinical correlation to fully assess the translational potential of these findings. This study is based on bioinformatics approach, has several limitations. It relies solely on publicly available transcriptomic data without experimental validation, which is necessary to confirm the biological roles of the identified DE-FRGs in PDAC. The relatively small sample size and absence of an independent validation cohort may limit the generalizability of the findings. Additionally, we did not construct a gene expression-based prognostic scoring model, which could enhance clinical relevance. Lastly, the predicted drug-gene interactions require further pharmacological validation to assess their therapeutic potential in PDAC.
Conclusions
The current study systematically the expression of biomarkers for ferroptosis in PDAC and determines CYGB, EGR1, NTS, ATP6V1G2, SLC2A3, and SLC2A6 as the important factors for prognosis. These findings do not only advance our molecular insights into PDAC but also create a foundation for the invention of new ferroptosis-targeted therapies. Combination of the existing gaps in the research by conducting more elaborate experiments, investigation of the relation between various types of cell death, and tumor heterogeneity will be essential for the application of the findings into practice. It is therefore envisaged that, as further studies are conducted in the field of ferroptosis, it will pave way for identification of other intervention points that could be useful in managing PDAC as well as other malignancies.
V体育平台登录 - Acknowledgments
The authors are grateful to the Abdul Wali Khan University for providing the Lab and essential systems for analysis.
VSports app下载 - Footnote
Peer Review File: Available at https://apc.amegroups.com/article/view/10.21037/apc-25-2/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://apc.amegroups.com/article/view/10.21037/apc-25-2/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. As this study was conducted exclusively using publicly available transcriptomic datasets (GEO: GSE235516 and GSE268948) and did not involve any direct human or animal participants, ethical approval and informed consent were not required. Therefore, the ethical statement is not applicable in this context.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pitman MB, Centeno BA, Reid MD, et al. The World Health Organization Reporting System for Pancreaticobiliary Cytopathology. Acta Cytol 2023;67:304-20. [Crossref] [PubMed]
- Caban M, Małecka-Wojciesko E. Gaps and Opportunities in the Diagnosis and Treatment of Pancreatic Cancer. Cancers (Basel) 2023;15:5577. [Crossref] [PubMed]
- Niger M, Prisciandaro M, Antista M, et al. One size does not fit all for pancreatic cancers: A review on rare histologies and therapeutic approaches. World J Gastrointest Oncol 2020;12:833-49. [Crossref] [PubMed]
- Gupta DS, Gupta DS, Shetty SR. A Recent Overview of the Growing Applications of Biosimilars in Pancreatic Cancer Management: Current Picture and Future Perspectives. In: Bhatt S, Dureja H, Gunvantbhai Patel S, et al., eds. Biosimilars for Cancer Treatment. Springer, Singapore; 2024.
- Elsherif SB, Virarkar M, Javadi S, et al. Pancreatitis and PDAC: association and differentiation. Abdom Radiol (NY) 2020;45:1324-37. [Crossref] [PubMed]
- Słodkowski M, Wroński M, Karkocha D, et al. Current Approaches for the Curative-Intent Surgical Treatment of Pancreatic Ductal Adenocarcinoma. Cancers (Basel) 2023;15:2584. [Crossref] [PubMed]
- Li C, Yin X, Liu Z, et al. Emerging Potential Mechanism and Therapeutic Target of Ferroptosis in PDAC: A Promising Future. Int J Mol Sci 2022;23:15031. [Crossref] [PubMed]
- Yu H, Guo P, Xie X, et al. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J Cell Mol Med 2017;21:648-57. [Crossref (V体育安卓版)] [PubMed]
- Li FJ, Long HZ, Zhou ZW, et al. System Xc-/GSH/GPX4 axis: An important antioxidant system for the ferroptosis in drug-resistant solid tumor therapy. Front Pharmacol 2022;13:910292. [Crossref] [PubMed]
- Yang J, Dai X, Xu H, et al. Regulation of Ferroptosis by Amino Acid Metabolism in Cancer. Int J Biol Sci 2022;18:1695-705. [Crossref (VSports最新版本)] [PubMed]
- Wang L, Chen X, Yan C. Ferroptosis: An emerging therapeutic opportunity for cancer. Genes Dis 2022;9:334-46. ["VSports app下载" Crossref] [PubMed]
- Jin W, Zhuang X, Lin Y, et al. Integrating ferroptosis-related genes (FRGs) and prognostic models to enhance UCEC outcome prediction and therapeutic insights. J Appl Genet 2023;64:723-35. [Crossref (VSports app下载)] [PubMed]
- Lopez-Blazquez C, Lacalle-Gonzalez C, Sanz-Criado L, et al. Iron-Dependent Cell Death: A New Treatment Approach against Pancreatic Ductal Adenocarcinoma. Int J Mol Sci 2023;24:14979. [Crossref] [PubMed]
- Yi S, Zhang C, Li M, Wang J. Construction of a Novel Diagnostic Model Based on Ferroptosis-Related Genes for Hepatocellular Carcinoma Using Machine and Deep Learning Methods. J Oncol 2023;2023:1624580. [Crossref] [PubMed]
- Li N, Jiang W, Wang W, et al. Ferroptosis and its emerging roles in cardiovascular diseases. Pharmacol Res 2021;166:105466. [Crossref] [PubMed]
- Han C, Liu Y, Dai R, et al. Ferroptosis and Its Potential Role in Human Diseases. Front Pharmacol 2020;11:239. [Crossref] [PubMed]
- Principe DR, Underwood PW, Korc M, et al. The Current Treatment Paradigm for Pancreatic Ductal Adenocarcinoma and Barriers to Therapeutic Efficacy. Front Oncol 2021;11:688377. ["VSports最新版本" Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Zhang H, Sun Y, Wang Z, et al. ZDHHC20-mediated S-palmitoylation of YTHDF3 stabilizes MYC mRNA to promote pancreatic cancer progression. Nat Commun 2024;15:4642. [V体育安卓版 - Crossref] [PubMed]
- Barrett T, Suzek TO, Troup DB, et al. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res 2005;33:D562-6. [Crossref] [PubMed]
- Modi A, Vai S, Caramelli D, et al. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Methods Mol Biol 2021;2242:15-42. [Crossref] [PubMed]
- Paijmans JLA, Baleka S, Henneberger K, et al. Sequencing single-stranded libraries on the Illumina NextSeq 500 platform. arXiv Prepr arXiv171111004. 2017.
- Varet H, Brillet-Guéguen L, Coppée JY, et al. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS One 2016;11:e0157022. [Crossref] [PubMed]
- Gu Z. Complex heatmap visualization. Imeta 2022;1:e43. [Crossref] [PubMed]
- Aiman S, Ahmad A, Khan AA, et al. Vaccinomics-based next-generation multi-epitope chimeric vaccine models prediction against Leishmania tropica - a hierarchical subtractive proteomics and immunoinformatics approach. Front Immunol 2023;14:1259612. [Crossref (V体育官网入口)] [PubMed]
- Berkman SJ, Roscoe EM, Bourret JC. Comparing self-directed methods for training staff to create graphs using Graphpad Prism. J Appl Behav Anal 2019;52:188-204. [Crossref] [PubMed]
- Manzoor U, Ali A, Ali SL, et al. Mutational screening of GDAP1 in dysphonia associated with Charcot-Marie-Tooth disease: clinical insights and phenotypic effects. J Genet Eng Biotechnol 2023;21:119. [Crossref] [PubMed]
- Zhou T, Yao J, Liu Z. Gene ontology, enrichment analysis, and pathway analysis. In: Liu Z, editor. Bioinformatics in Aquaculture 2017;150-68.
- Ali SL, Ali A, Alamri A, et al. Genomic annotation for vaccine target identification and immunoinformatics-guided multi-epitope-based vaccine design against Songling virus through screening its whole genome encoded proteins. Front Immunol 2023;14:1284366. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Pages H, Carlson M, Falcon S, et al. Package ‘AnnotationDbi.’ Bioconductor Packag Maint; 2017.
- Song Y, Liu M, Jia WP, et al. The association between nutritional status and functional limitations among centenarians: a cross-sectional study. BMC Geriatr 2021;21:376. [Crossref] [PubMed]
- Villanueva RAM, Chen ZJ. ggplot2: elegant graphics for data analysis. Taylor & Francis; 2019.
- Zhuang L, Ali A, Yang L, et al. Leveraging computer-aided design and artificial intelligence to develop a next-generation multi-epitope tuberculosis vaccine candidate. Infect Med (Beijing) 2024;3:100148. ["V体育安卓版" Crossref] [PubMed]
- Zhuang L, Zhao Y, Yang L, et al. Harnessing bioinformatics for the development of a promising multi-epitope vaccine against tuberculosis: The ZL9810L vaccine. Decod Infect Transm 2024;2:100026.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Ali SL, Ali A, Ullah W, et al. Promising vaccine models against astrovirus MLB2 using integrated vaccinomics and immunoinformatics approaches. Mol Syst Des Eng 2024;9:1285-99.
- Ali A, Ali SL, Alamri A, et al. Multi-epitope-based vaccine models prioritization against Astrovirus MLB1 using immunoinformatics and reverse vaccinology approaches. J Genet Eng Biotechnol 2025;23:100451. [Crossref] [PubMed]
- Luqman Ali S, Ali A, Ullah W, et al. Exploring advanced genomic and immunoinformatics techniques for identifying drug and vaccine targets against SARS-CoV-2. J Genet Eng Biotechnol 2024;22:100439. [Crossref] [PubMed]
- Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Ali A, Luqman Ali S. A Stable mRNA-Based Novel Multi-Epitope Vaccine Designs Against Infectious Heartland Virus by Integrated Immunoinformatics and Reverse Vaccinology Approaches. Viral Immunol 2025;38:73-87. [Crossref (VSports)] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Zhu J, Ye Z, Zhang Z, et al. AI-Integrated 3D imaging and modelling for hip morphology assessment in athletes. Comput Methods Biomech Biomed Engin 2025; Epub ahead of print. [Crossref]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Ali A, Ali SL, Ullah W, et al. Gene Expression Profiling Identifies CAV1, CD44, and TFRC as Potential Diagnostic Markers and Therapeutic Targets for Multiple Myeloma. Cell Biochem Biophys 2025; Epub ahead of print. [Crossref (V体育平台登录)]
- Khatrawi EM, Ali SL, Ali SY, et al. Designing a multi-epitope vaccine targeting UPF0721 of meningitis-causing Salmonella enterica serovar Typhimurium strain L-4126 by utilizing immuno-informatics and in silico approaches. Mol Syst Des Eng 2025;10:549-66.
- Keim DA, Mansmann F, Schneidewind J, et al. Visual analytics: Scope and challenges. In: Visual data mining: Theory, techniques and tools for visual analytics. Springer; 2008. p. 76-90.
- Ranstam J, Cook JA. Kaplan-Meier curve. Br J Surg 2017;104:442. [Crossref] [PubMed]
- Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90-7. [Crossref] [PubMed]
- Li C, Li X, Deng Z, et al. EGR1 and EGR2 positively regulate plant ABA signaling by modulating the phosphorylation of SnRK2.2. New Phytol 2024;241:1492-509. [Crossref] [PubMed]
- Chai F, Zhang J, Fu T, et al. Identification of SLC2A3 as a prognostic indicator correlated with the NF-κB/EMT axis and immune response in head and neck squamous cell carcinoma. Channels (Austin) 2023;17:2208928. [Crossref] [PubMed]
- Michalak N, Małecka-Wojciesko E. Modifiable Pancreatic Ductal Adenocarcinoma (PDAC) Risk Factors. J Clin Med 2023;12:4318. [Crossref] [PubMed]
- Clementine R, Leo M, Sandra G, et al. Management of pancreatic ductal adenocarcinoma (PDAC): Progress in the past decade and challenges for the future. Cancer Rep Rev 2019;3:1-12.
- Jentzsch V, Davis JAA, Djamgoz MBA. Pancreatic Cancer (PDAC): Introduction of Evidence-Based Complementary Measures into Integrative Clinical Management. Cancers (Basel) 2020;12:3096. [Crossref (V体育安卓版)] [PubMed]
- Lee DH, Imran M, Choi JH, et al. CDK4/6 inhibitors induce breast cancer senescence with enhanced anti-tumor immunogenic properties compared with DNA-damaging agents. Mol Oncol 2024;18:216-32. [Crossref] [PubMed]
- Ali A, Ali SL, Omneya A. A Comprehensive Methodological Review of Major Developments in Bioinformatics Pipelines for Transcriptomic Data Analysis. Nov Biomed 2025;13:46-60.
- Ali A, Manzoor U, Ali SL, et al. Analysis of the capability of IgG antibodies and receptors with their relationships to food tolerance and autoimmune disorders. Int J Nat Med Heal Sci 2023;3:25-32.
- ALi A. Currently trending and futuristic biological modalities in the management of different types of diabetes: A comprehensive review. J Popul Ther Clin Pharmacol 2023;30:2948-70.
- Xiao F, Wang K, Cheng Q. Porphyry magma cooling and crystallization control of mineralization: Insights from the dynamic numerical modeling. Ore Geol Rev. 2024;166:105956.
- Cannon M, Stevenson J, Stahl K, et al. DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res 2024;52:D1227-35. [Crossref] [PubMed]
- Khatrawi EM, Luqman Ali S, Ali SY, et al. Robust Multiepitope Vaccine from Glycoproteins Against Human Metapneumovirus Genotypes A2a, A2b, and A2c by Utilizing Immunoinformatics and Reverse Vaccinology Approaches. Viral Immunol 2025;38:157-71. [Crossref] [PubMed]
- Suresh S, Freedman A, Adams M, et al. Placental histology for targeted risk assessment of recurrent spontaneous preterm birth. Am J Obstet Gynecol 2024;230:452.e1-452.e11. [Crossref] [PubMed]
Cite this article as: Ali SL, Ali A, Khan A. Identification and assessment of ferroptosis-related genes and their implication as therapeutic agents for pancreatic ductal adenocarcinoma. Ann Pancreat Cancer 2025;8:6.


 "VSports"
"VSports"