Identification of diverse lipid droplet targeting motifs in the PNPLA family of triglyceride lipases
- PMID: 23741432
- PMCID: PMC3669214
- DOI: 10.1371/journal.pone.0064950
V体育安卓版 - Identification of diverse lipid droplet targeting motifs in the PNPLA family of triglyceride lipases
Abstract
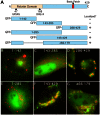
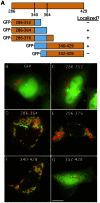
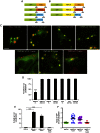
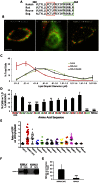
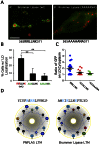
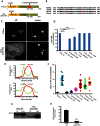
Members of the Patatin-like Phospholipase Domain containing Protein A (PNPLA) family play key roles in triglyceride hydrolysis, energy metabolism, and lipid droplet (LD) homoeostasis. Here we report the identification of two distinct LD targeting motifs (LTM) for PNPLA family members. Transient transfection of truncated versions of human adipose triglyceride lipase (ATGL, also known as PNPLA2), PNPLA3/adiponutrin, or PNPLA5 (GS2-like) fused to GFP revealed that the C-terminal third of these proteins contains sequences that are sufficient for targeting to LDs. Furthermore, fusing the C-termini of PNPLA3 or PNPLA5 confers LD localization to PNPLA4, which is otherwise cytoplasmic. Analyses of additional mutants in ATGL, PNPLA5, and Brummer Lipase, the Drosophila homolog of mammalian ATGL, identified two different types of LTMs. The first type, in PNPLA5 and Brummer lipase, is a set of loosely conserved basic residues, while the second type, in ATGL, is contained within a stretch of hydrophobic residues. These results show that even closely related members of the PNPLA family employ different molecular motifs to associate with LDs. VSports手机版.
Conflict of interest statement
Figures
References
-
- Brasaemle DL, Wolins NE (2012) Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem 287: 2273–2279. - VSports最新版本 - PMC - PubMed
Publication types
MeSH terms
- "V体育ios版" Actions
- "VSports手机版" Actions
- "V体育ios版" Actions
- Actions (V体育2025版)
- "VSports在线直播" Actions
- Actions (VSports手机版)
- V体育安卓版 - Actions
- "VSports" Actions
Substances
- "V体育安卓版" Actions
- V体育安卓版 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
"VSports手机版" Research Materials

