Fructose-1,6-bisphosphatase opposes renal carcinoma progression
- PMID: 25043030
- PMCID: PMC4162811
- DOI: 10.1038/nature13557
VSports最新版本 - Fructose-1,6-bisphosphatase opposes renal carcinoma progression
Abstract
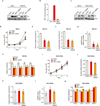
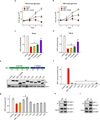
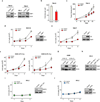
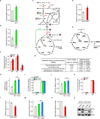
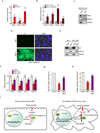
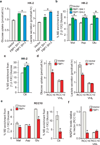
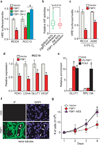
Clear cell renal cell carcinoma (ccRCC), the most common form of kidney cancer, is characterized by elevated glycogen levels and fat deposition. These consistent metabolic alterations are associated with normoxic stabilization of hypoxia-inducible factors (HIFs) secondary to von Hippel-Lindau (VHL) mutations that occur in over 90% of ccRCC tumours. However, kidney-specific VHL deletion in mice fails to elicit ccRCC-specific metabolic phenotypes and tumour formation, suggesting that additional mechanisms are essential. Recent large-scale sequencing analyses revealed the loss of several chromatin remodelling enzymes in a subset of ccRCC (these included polybromo-1, SET domain containing 2 and BRCA1-associated protein-1, among others), indicating that epigenetic perturbations are probably important contributors to the natural history of this disease. Here we used an integrative approach comprising pan-metabolomic profiling and metabolic gene set analysis and determined that the gluconeogenic enzyme fructose-1,6-bisphosphatase 1 (FBP1) is uniformly depleted in over six hundred ccRCC tumours examined VSports手机版. Notably, the human FBP1 locus resides on chromosome 9q22, the loss of which is associated with poor prognosis for ccRCC patients. Our data further indicate that FBP1 inhibits ccRCC progression through two distinct mechanisms. First, FBP1 antagonizes glycolytic flux in renal tubular epithelial cells, the presumptive ccRCC cell of origin, thereby inhibiting a potential Warburg effect. Second, in pVHL (the protein encoded by the VHL gene)-deficient ccRCC cells, FBP1 restrains cell proliferation, glycolysis and the pentose phosphate pathway in a catalytic-activity-independent manner, by inhibiting nuclear HIF function via direct interaction with the HIF inhibitory domain. This unique dual function of the FBP1 protein explains its ubiquitous loss in ccRCC, distinguishing FBP1 from previously identified tumour suppressors that are not consistently mutated in all tumours. .
Figures







Comment in
-
Kidney cancer: FBP1 depletion feeds ccRCC.Nat Rev Urol. 2014 Sep;11(9):482. doi: 10.1038/nrurol.2014.200. Epub 2014 Aug 5. Nat Rev Urol. 2014. PMID: 25091010 No abstract available.
-
Tumorigenesis: FBP1 is suppressed in kidney tumours.Nat Rev Cancer. 2014 Sep;14(9):575. doi: 10.1038/nrc3810. Nat Rev Cancer. 2014. PMID: 25145476 No abstract available.
-
V体育ios版 - Re: Fructose-1,6-bisphosphatase opposes renal carcinoma progression.J Urol. 2015 May;193(5):1725-6. doi: 10.1016/j.juro.2015.02.022. Epub 2015 Feb 10. J Urol. 2015. PMID: 25895807 No abstract available.
"VSports手机版" References
-
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. - "V体育安卓版" PubMed
-
- Valera VA, Merino MJ. Misdiagnosis of clear cell renal cell carcinoma. Nat Rev Urol. 2011;8:321–333. - VSports app下载 - PubMed
-
- Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. - VSports手机版 - PMC - PubMed
Publication types
- "V体育ios版" Actions
MeSH terms
- "V体育平台登录" Actions
- VSports最新版本 - Actions
- Actions (V体育ios版)
- Actions (V体育ios版)
- Actions (V体育安卓版)
- Actions (V体育2025版)
- VSports在线直播 - Actions
- "V体育安卓版" Actions
- Actions (V体育平台登录)
- "VSports app下载" Actions
- "VSports注册入口" Actions
- Actions (V体育安卓版)
"V体育2025版" Substances
- "VSports手机版" Actions
Grants and funding
LinkOut - more resources
"V体育官网" Full Text Sources
Other Literature Sources
"VSports注册入口" Medical
Molecular Biology Databases
Research Materials
Miscellaneous

