Modulation of macrophage inflammatory function through selective inhibition of the epigenetic reader protein SP140 (VSports最新版本)
- PMID: 35986286
- PMCID: PMC9392322
- DOI: 10.1186/s12915-022-01380-6
V体育官网 - Modulation of macrophage inflammatory function through selective inhibition of the epigenetic reader protein SP140
Abstract
Background: SP140 is a bromodomain-containing protein expressed predominantly in immune cells. Genetic polymorphisms and epigenetic modifications in the SP140 locus have been linked to Crohn's disease (CD), suggesting a role in inflammation VSports手机版. .
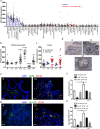
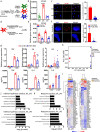
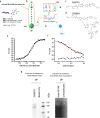
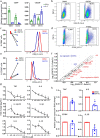
Results: We report the development of the first small molecule SP140 inhibitor (GSK761) and utilize this to elucidate SP140 function in macrophages. We show that SP140 is highly expressed in CD mucosal macrophages and in in vitro-generated inflammatory macrophages V体育安卓版. SP140 inhibition through GSK761 reduced monocyte-to-inflammatory macrophage differentiation and lipopolysaccharide (LPS)-induced inflammatory activation, while inducing the generation of CD206+ regulatory macrophages that were shown to associate with a therapeutic response to anti-TNF in CD patients. SP140 preferentially occupies transcriptional start sites in inflammatory macrophages, with enrichment at gene loci encoding pro-inflammatory cytokines/chemokines and inflammatory pathways. GSK761 specifically reduces SP140 chromatin binding and thereby expression of SP140-regulated genes. GSK761 inhibits the expression of cytokines, including TNF, by CD14+ macrophages isolated from CD intestinal mucosa. .
Conclusions: This study identifies SP140 as a druggable epigenetic therapeutic target for CD V体育ios版. .
Keywords: Crohn's disease; Macrophage; SP140 VSports最新版本. .
© 2022. The Author(s).
"VSports注册入口" Conflict of interest statement
PDC, AYFLY, AS, SH, RPB, KG, GY, HPK, AC, AP, LAS, DJM, LL, TGH, RJW, CEB, LAH, GB, UK, NH, JRP, RKP, NRH, and DFT were employed by GSK at the time of conducting this study. MG, JK, ILH, DAZ, OW, TBMH, AATV, JVL, PH, MPJW, and WJDJ were employed by Amsterdam University Medical Centers at the time of conducting this study V体育平台登录. GSK has a patent EP2643462B1 related to the therapeutic use of SP140 inhibitors.
Figures


"VSports app下载" References
-
- Papadakis KA, Shaye OA, Vasiliauskas EA, Ippoliti A, Dubinsky MC, Birt J, et al. Safety and efficacy of adalimumab (D2E7) in Crohn's disease patients with an attenuated response to infliximab. Am J Gastroenterol. 2005;100(1):75–79. doi: 10.1111/j.1572-0241.2005.40647.x. - "VSports注册入口" DOI - PubMed
Publication types
- Actions (VSports注册入口)
MeSH terms
- "VSports最新版本" Actions
- VSports在线直播 - Actions
- "VSports最新版本" Actions
- V体育安卓版 - Actions
- V体育官网 - Actions
- Actions (V体育官网)
- VSports在线直播 - Actions
Substances
- "V体育平台登录" Actions
- "VSports在线直播" Actions
LinkOut - more resources
Full Text Sources
Medical (VSports注册入口)
Research Materials

