HER3 activation contributes toward the emergence of ALK inhibitor-tolerant cells in ALK-rearranged lung cancer with mesenchymal features (VSports app下载)
- PMID: 35042943
- PMCID: PMC8766605
- DOI: 10.1038/s41698-021-00250-8 (V体育安卓版)
HER3 activation contributes toward the emergence of ALK inhibitor-tolerant cells in ALK-rearranged lung cancer with mesenchymal features
Abstract
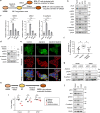
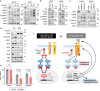
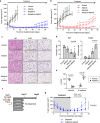
Anaplastic lymphoma kinase-tyrosine kinase inhibitors (ALK-TKIs) have shown dramatic efficacy in patients with ALK-rearranged lung cancer; however, complete response in these patients is rare VSports手机版. Here, we investigated the molecular mechanisms underlying the emergence and maintenance of drug-tolerant cells in ALK-rearranged lung cancer. Cell based-assays demonstrated that HER3 activation and mesenchymal-to-epithelial transition, mediated through ZEB1 proteins, help maintain cell survival and induce the emergence of ALK-TKI-tolerant cells. Compared with ALK-TKIs alone, cotreatment with pan-HER inhibitor afatinib and ALK-TKIs prevented tumor regrowth, leading to the eradication of tumors in ALK-rearranged tumors with mesenchymal features. Moreover, pre-treatment vimentin expression in clinical specimens obtained from patients with ALK-rearranged lung cancer was associated with poor ALK-TKI treatment outcomes. These results demonstrated that HER3 activation plays a pivotal role in the emergence of ALK-TKI-tolerant cells. Furthermore, the inhibition of HER3 signals combined with ALK-TKIs dramatically improves treatment outcomes for ALK-rearranged lung cancer with mesenchymal features. .
© 2022. The Author(s).
"VSports手机版" Conflict of interest statement
T. Y. received commercial research grants from Pfizer, Ono Pharmaceutical, Chugai Pharmaceutical, and Takeda Pharmaceutical Company Limited. J. U. received research grants from Eli Lilly Japan K. K. , AstraZeneca K. K. , and Boehringer Ingelheim Japan. T. S. received research grants from Otsuka Pharmaceutical and Oncolys BioPharma, and a patent fee from JT Pharmaceutical. S. Y. obtained commercial research grants from Chugai Pharmaceutical and Boehringer-Ingelheim and has received speaking honoraria from Chugai Pharmaceutical, Boehringer-Ingelheim, Novartis, and Pfizer. R. K. received research grants from Chugai Pharmaceutical, Takeda Pharmaceutical Company Limited, Toppan Printing, and Daiichi-Sankyo V体育安卓版. K. T. received research grants from Chugai-Roche and Ono Pharmaceutical and personal fees from AstraZeneca, Chugai-Roche, MSD-Merck, Eli Lilly, Boehringer-Ingelheim, and Daiichi-Sankyo. The remaining authors declare no competing interests.
Figures


References
-
- Miller KD, et al. Cancer statistics for hispanics/latinos, 2018. CA Cancer J. Clin. 2018;68:425–445. - "VSports app下载" PubMed
-
- Govindan R, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol. 2006;24:4539–4544. - VSports - PubMed
-
- Recondo G, Facchinetti F, Olaussen KA, Besse B, Friboulet L. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat. Rev. Clin. Oncol. 2018;15:694–708. - PubMed
-
- Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. - PubMed
Grants and funding
- 19K08608/MEXT | Japan Society for the Promotion of Science (JSPS)
- V体育平台登录 - 20K22820/MEXT | Japan Society for the Promotion of Science (JSPS)
- 19H03524/MEXT | Japan Society for the Promotion of Science (JSPS) (VSports最新版本)
- 2018A18/Japan Research Foundation for Clinical Pharmacology
- JP20cm0106203h0005/Japan Agency for Medical Research and Development (AMED)
LinkOut - more resources
"V体育官网入口" Full Text Sources
Molecular Biology Databases
Research Materials

