Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk (VSports注册入口)
- PMID: 31992511
- PMCID: "V体育安卓版" PMC7300386
- DOI: 10.1016/j.molmet.2019.12.005
Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk
VSports app下载 - Abstract
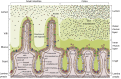
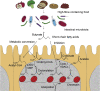
Background: The microbiota in the human gut are an important component of normal physiology that has co-evolved from the earliest multicellular organisms. Therefore, it is unsurprising that there is intimate crosstalk between the microbial world in the gut and the host VSports手机版. Genome regulation through microbiota-host interactions not only affects the host's immunity, but also metabolic health and resilience against cancer. Chromatin dynamics of the host epithelium involving histone modifications and other facets of the epigenetic machinery play an important role in this process. .
Scope of review: This review discusses recent findings relevant to how chromatin dynamics shape the crosstalk between the microbiota and its host, with a special focus on the role of histone modifications V体育安卓版. .
Major conclusions: Host-microbiome interactions are important evolutionary drivers and are thus expected to be hardwired into and mould the epigenetic machinery in multicellular organisms. Microbial-derived short-chain fatty acids (SCFA) are dominant determinants of microbiome-host interactions, and the inhibition of histone deacetylases (HDACs) by SCFA is a key mechanism in this process. The discovery of alternative histone acylations, such as crotonylation, in addition to the canonical histone acetylation reveals a new layer of complexity in this crosstalk V体育ios版. .
Keywords: Acylations; Chromatin; Crotonylation; Histone modifications; Microbiome; Microbiota. VSports最新版本.
Copyright © 2019 The Authors V体育平台登录. Published by Elsevier GmbH. All rights reserved. .
Figures

References
-
- Castillo J., López-Rodas G., Franco L. Histone post-translational modifications and nucleosome organisation in transcriptional regulation: some open questions. Advances in Experimental Medicine and Biology. 2017;966:65–92. - PubMed
-
- Suganuma T., Workman J.L. Signals and combinatorial functions of histone modifications. Annual Review of Biochemistry. 2011;80:473–499. - "VSports注册入口" PubMed
-
- Yun M., Wu J., Workman J.L., Li B. Readers of histone modifications. Cell Research. 2011;21:564–578. - PMC (V体育2025版) - PubMed
-
- Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Experimental and Molecular Medicine. 2017;49:e324. - "VSports注册入口" PMC - PubMed
-
- Varga-Weisz P.D. Chromatin remodeling: a collaborative effort. Nature Structural and Molecular Biology. 2014;21:14–16. - PubMed
"VSports手机版" Publication types
MeSH terms
- "V体育2025版" Actions
- VSports手机版 - Actions
- V体育官网 - Actions
- Actions (V体育ios版)
- VSports app下载 - Actions
Substances
- "VSports手机版" Actions
- "V体育2025版" Actions
Grants and funding
VSports注册入口 - LinkOut - more resources
Full Text Sources
Research Materials

