Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1 (V体育官网)
- PMID: 31369196
- PMCID: PMC6887590
- DOI: "VSports注册入口" 10.1002/1878-0261.12555
Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1
Abstract (VSports)
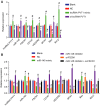
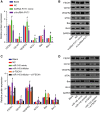
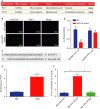
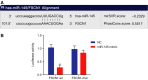
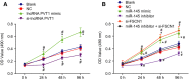
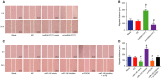
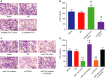
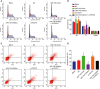
Accumulating evidence has established that long noncoding RNA (lncRNA) plasmacytoma variant translocation 1 (PVT1) is a tumor regulator in many cancers. Here, we aimed to investigate the possible function of lncRNA PVT1 in esophageal carcinoma (EC) via targeting of microRNA-145 (miR-145). Initially, microarray-based gene expression profiling of EC was employed to identify differentially expressed genes. Moreover, the expression of lncRNA PVT1 was examined and the cell line presenting with the highest level of lncRNA PVT1 expression was selected for subsequent experiments. We then proceeded to examine interaction among lncRNA PVT1, FSCN1, and miR-145. The effect of lncRNA PVT1 on viability, migration, invasion, apoptosis, and tumorigenesis of transfected cells was examined with gain-of-function and loss-of-function experiments. We observed that lncRNA PVT1 was robustly induced in EC. lncRNA PVT1 could bind to miR-145 and regulate its expression, and FSCN1 is a target gene of miR-145 VSports手机版. Overexpression of miR-145 or silencing of lncRNA PVT1 was revealed to suppress cell viability, migration, and invasion abilities, while also stimulating cell apoptosis. Furthermore, our in vivo results showed that overexpression of miR-145 or silencing of lncRNA PVT1 resulted in decreased tumor growth in nude mice. In conclusion, our research reveals that down-regulation of lncRNA PVT1 could potentially promote expression of miR-145 to repress cell migration and invasion, and promote cell apoptosis through the inhibition of FSCN1. This highlights the potential of lncRNA PVT1 as a therapeutic target for EC treatment. .
Keywords: FSCN1; apoptosis; esophageal carcinoma; long noncoding RNA; microRNA-145; plasmacytoma variant translocation 1 V体育安卓版. .
© 2019 The Authors. Published by FEBS Press and John Wiley & Sons Ltd V体育ios版. .
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chang L, Yuan Z, Shi H, Bian Y and Guo R (2017) miR‐145 targets the SOX11 3′UTR to suppress endometrial cancer growth. Am J Cancer Res 7, 2305–2317. - "VSports最新版本" PMC - PubMed
-
- Chen X, Gong J, Zeng H, Chen N, Huang R, Huang Y, Nie L, Xu M, Xia J, Zhao F et al (2010) MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res 70, 2728–2738. - PubMed
Publication types
"VSports在线直播" MeSH terms
- Actions (V体育官网入口)
- Actions (V体育官网入口)
- "VSports" Actions
- Actions (VSports注册入口)
- "V体育平台登录" Actions
- V体育ios版 - Actions
- "VSports app下载" Actions
- "VSports最新版本" Actions
- Actions (VSports)
- VSports注册入口 - Actions
- Actions (V体育2025版)
- "V体育安卓版" Actions
- Actions (V体育ios版)
- "V体育2025版" Actions
- VSports注册入口 - Actions
- VSports app下载 - Actions
- "VSports手机版" Actions
Substances
- "VSports" Actions
- "VSports手机版" Actions
- "VSports" Actions
- VSports最新版本 - Actions
- Actions (VSports最新版本)
- Actions (VSports)
- "V体育官网" Actions
"VSports app下载" LinkOut - more resources
Full Text Sources
Medical
V体育安卓版 - Miscellaneous

