Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel
- PMID: 30816954
- PMCID: PMC6512308
- DOI: 10.1001/jamaoncol.2018.7098
Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel
"VSports注册入口" Abstract
Importance: Tumor mutational burden (TMB), as measured by whole-exome sequencing (WES) or a cancer gene panel (CGP), is associated with immunotherapy responses. However, whether TMB estimated by circulating tumor DNA in blood (bTMB) is associated with clinical outcomes of immunotherapy remains to be explored VSports手机版. .
Objectives: To explore the optimal gene panel size and algorithm to design a CGP for TMB estimation, evaluate the panel reliability, and further validate the feasibility of bTMB as a clinical actionable biomarker for immunotherapy. V体育安卓版.
Design, setting, and participants: In this cohort study, a CGP named NCC-GP150 was designed and virtually validated using The Cancer Genome Atlas database V体育ios版. The correlation between bTMB estimated by NCC-GP150 and tissue TMB (tTMB) measured by WES was evaluated in matched blood and tissue samples from 48 patients with advanced NSCLC. An independent cohort of 50 patients with advanced NSCLC was used to identify the utility of bTMB estimated by NCC-GP150 in distinguishing patients who would benefit from anti-programmed cell death 1 (anti-PD-1) and anti-programmed cell death ligand 1 (anti-PD-L1) therapy. The study was performed from July 19, 2016, to April 20, 2018. .
Main outcomes and measures: Assessment of the Spearman correlation coefficient between bTMB estimated by NCC-GP150 and tTMB calculated by WES VSports最新版本. Evaluation of the association of bTMB level with progression-free survival and response to anti-PD-1 and anti-PD-L1 therapy. .
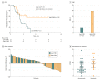
Results: This study used 2 independent cohorts of patients with NSCLC (cohort 1: 48 patients; mean [SD] age, 60 [13] years; 15 [31. 2%] female; cohort 2: 50 patients; mean [SD] age, 58 [8] years; 15 [30. 0%] female). A CGP, including 150 genes, demonstrated stable correlations with WES for TMB estimation (median r2 = 0. 91; interquartile range, 0. 89-0. 92), especially when synonymous mutations were included (median r2 = 0. 92; interquartile range, 0. 91-0. 93), whereas TMB estimated by the NCC-GP150 panel found higher correlations with TMB estimated by WES than most of the randomly sampled 150-gene panels. Blood TMB estimated by NCC-GP150 correlated well with the matched tTMB calculated by WES (Spearman correlation = 0 V体育平台登录. 62). In the anti-PD-1 and anti-PD-L1 treatment cohort, a bTMB of 6 or higher was associated with superior progression-free survival (hazard ratio, 0. 39; 95% CI, 0. 18-0. 84; log-rank P = . 01) and objective response rates (bTMB ≥6: 39. 3%; 95% CI, 23. 9%-56. 5%; bTMB <6: 9. 1%; 95% CI, 1. 6%-25. 9%; P = . 02). .
Conclusions and relevance: The findings suggest that established NCC-GP150 with an optimized gene panel size and algorithm is feasible for bTMB estimation, which may serve as a potential biomarker of clinical benefit in patients with NSCLC treated with anti-PD-1 and anti-PD-L1 agents. VSports注册入口.
Conflict of interest statement
Conflict of Interest Disclosures: Drs M. Han, Dong, Lu, Cao, Li, D V体育官网入口. Wang, Chen, Xu, X. Zhao, G. Wang, Y. Bai, J. Zhao, Z. Zhao, Zhang, Xiong, and Cai are employees of 3D Medicines Inc. No other disclosures were reported.
Figures

Comment in
-
V体育官网入口 - bTMB is a promising predictive biomarker.Nat Rev Clin Oncol. 2019 Jul;16(7):403. doi: 10.1038/s41571-019-0202-8. Nat Rev Clin Oncol. 2019. PMID: 30874612 No abstract available.
-
V体育官网入口 - Blood-based tumor mutation burden: continued progress toward personalizing immunotherapy in non-small cell lung cancer.J Thorac Dis. 2019 Jun;11(6):2208-2211. doi: 10.21037/jtd.2019.05.68. J Thorac Dis. 2019. PMID: 31372254 Free PMC article. No abstract available.
-
V体育平台登录 - Blood tumor mutational burden: are we ready for clinical implementation?J Thorac Dis. 2019 Sep;11(Suppl 15):S1906-S1908. doi: 10.21037/jtd.2019.07.60. J Thorac Dis. 2019. PMID: 31632782 Free PMC article. No abstract available.
References
-
- Snyder A, Makarov V, Merghoub T, et al. . Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189-2199. doi:10.1056/NEJMoa1406498 - "VSports最新版本" DOI - PMC - PubMed
-
- Van Allen EM, Miao D, Schilling B, et al. . Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207-211. doi:10.1126/science.aad0095 - V体育官网 - DOI - PMC - PubMed
MeSH terms
- "V体育安卓版" Actions
- V体育2025版 - Actions
- V体育2025版 - Actions
- "VSports" Actions
- Actions (V体育官网入口)
- Actions (VSports)
- VSports最新版本 - Actions
- Actions (V体育官网)
- "VSports注册入口" Actions
- V体育官网入口 - Actions
Substances
- "V体育ios版" Actions
- Actions (V体育官网)
LinkOut - more resources
Full Text Sources
Other Literature Sources (VSports手机版)
Medical
Research Materials
Miscellaneous

