Sex-chromosome dosage effects on gene expression in humans
- PMID: 29946024
- PMCID: "VSports最新版本" PMC6048519
- DOI: 10.1073/pnas.1802889115
Sex-chromosome dosage effects on gene expression in humans
Abstract
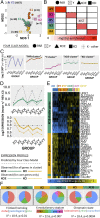
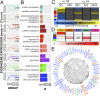
A fundamental question in the biology of sex differences has eluded direct study in humans: How does sex-chromosome dosage (SCD) shape genome function. To address this, we developed a systematic map of SCD effects on gene function by analyzing genome-wide expression data in humans with diverse sex-chromosome aneuploidies (XO, XXX, XXY, XYY, and XXYY) VSports手机版. For sex chromosomes, we demonstrate a pattern of obligate dosage sensitivity among evolutionarily preserved X-Y homologs and update prevailing theoretical models for SCD compensation by detecting X-linked genes that increase expression with decreasing X- and/or Y-chromosome dosage. We further show that SCD-sensitive sex-chromosome genes regulate specific coexpression networks of SCD-sensitive autosomal genes with critical cellular functions and a demonstrable potential to mediate previously documented SCD effects on disease. These gene coexpression results converge with analysis of transcription factor binding site enrichment and measures of gene expression in murine knockout models to spotlight the dosage-sensitive X-linked transcription factor ZFX as a key mediator of SCD effects on wider genome expression. Our findings characterize the effects of SCD broadly across the genome, with potential implications for human phenotypic variation. .
Keywords: Klinefelter syndrome; Turner syndrome; X-inactivation; sex chromosomes; sex differences V体育安卓版. .
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hughes JF, Rozen S. Genomics and genetics of human and primate y chromosomes. Annu Rev Genomics Hum Genet. 2012;13:83–108. - PubMed
-
- Bachtrog D, et al. Tree of Sex Consortium Sex determination: Why so many ways of doing it? PLoS Biol. 2014;12:e1001899. - VSports app下载 - PMC - PubMed
-
- Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci USA. 2010;107:3394–3399. - PMC (VSports注册入口) - PubMed
MeSH terms
- Actions (V体育ios版)
- Actions (V体育安卓版)
- V体育ios版 - Actions
- Actions (VSports手机版)
- "V体育平台登录" Actions
- "VSports app下载" Actions
- VSports - Actions
- "V体育平台登录" Actions
- Actions (VSports注册入口)
"VSports注册入口" Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

