Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins
- PMID: 29872532
- PMCID: PMC5981465
- DOI: 10.1038/s41438-018-0032-3 (V体育平台登录)
Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins
VSports手机版 - Abstract
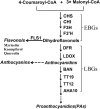
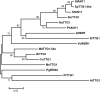
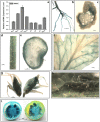
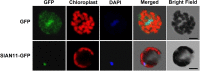
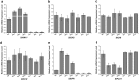
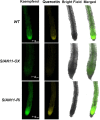
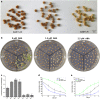
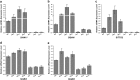
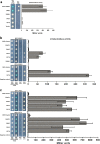
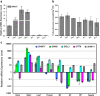
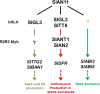
The flavonoid compounds are important secondary metabolites with versatile human nutritive benefits and fulfill a multitude of functions during plant growth and development VSports手机版. The abundance of different flavonoid compounds are finely tuned with species-specific pattern by a ternary MBW complex, which consists of a MYB, a bHLH, and a WD40 protein, but the essential role of SlAN11, which is a WD40 protein, is not fully understood in tomato until now. In this study, a tomato WD40 protein named as SlAN11 was characterized as an effective transcription regulator to promote plant anthocyanin and seed proanthocyanidin (PA) contents, with late flavonoid biosynthetic genes activated in 35S::SlAN11 transgenic lines, while the dihydroflavonol flow to the accumulation of flavonols or their glycosylated derivatives was reduced by repressing the expression of SlFLS in this SlAN11-overexpressed lines. The above changes were reversed in 35S::SlAN11-RNAi transgenic lines except remained levels of flavonol compounds and SlFLS expression. Interestingly, our data revealed that SlAN11 gene could affect seed dormancy by regulating the expressions of abscisic acid (ABA) signaling-related genes SlABI3 and SlABI5, and the sensitivity to ABA treatment in seed germination is conversely changed by SlAN11-overexpressed or -downregulated lines. Yeast two-hybrid assays demonstrated that SlAN11 interacted with bHLH but not with MYB proteins in the ternary MBW complex, whereas bHLH interacted with MYB in tomato. Our results indicated that low level of anthocyanins in tomato fruits, with low expression of bHLH (SlTT8) and MYB (SlANT1 and SlAN2) genes, remain unchanged upon modification of SlAN11 gene alone in the transgenic lines. These results suggest that the tomato WD40 protein SlAN11, coordinating with bHLH and MYB proteins, plays a crucial role in the fine adjustment of the flavonoid biosynthesis and seed dormancy in tomato. .
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

"VSports app下载" References
-
- Li S. Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014;9:e27522. doi: 10.4161/psb.27522. - DOI (V体育ios版) - PMC - PubMed
-
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant. Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. - "V体育官网入口" DOI - PubMed
-
- Ben-Simhon Z, et al. A pomegranate (punica granatum l.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta. 2011;234:865–881. doi: 10.1007/s00425-011-1438-4. - "V体育官网" DOI - PubMed
LinkOut - more resources (VSports手机版)
Full Text Sources
Other Literature Sources

