How cells ensure correct repair of DNA double-strand breaks
- PMID: 29414795
- PMCID: PMC6036189
- DOI: "V体育官网" 10.1074/jbc.TM118.000371
How cells ensure correct repair of DNA double-strand breaks (VSports注册入口)
VSports - Abstract
DNA double-strand breaks (DSBs) arise regularly in cells and when left unrepaired cause senescence or cell death. Homologous recombination (HR) and nonhomologous end-joining (NHEJ) are the two major DNA-repair pathways. Whereas HR allows faithful DSB repair and healthy cell growth, NHEJ has higher potential to contribute to mutations and malignancy. Many regulatory mechanisms influence which of these two pathways is used in DSB repair. These mechanisms depend on the cell cycle, post-translational modifications, and chromatin effects. Here, we summarize current research into these mechanisms, with a focus on mammalian cells, and also discuss repair by "alternative end-joining" and single-strand annealing. VSports手机版.
Keywords: BRCA1; DNA damage; DNA repair; chromatin; double-strand break; homologous recombination; nonhomologous end-joining; ubiquitylation (ubiquitination). V体育安卓版.
© 2018 Her and Bunting.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures (VSports手机版)
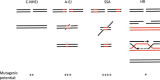
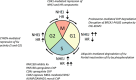
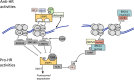
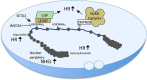
"V体育安卓版" References
-
- Helleday T., Eshtad S., and Nik-Zainal S. (2014) Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 15, 585–598 10.1038/nrg3729 - VSports手机版 - DOI - PMC - PubMed
-
- Kowalczykowski S. C. (2015) An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410 10.1101/cshperspect.a016410 - "VSports app下载" DOI - PMC - PubMed
-
- Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A. A., Krempler A., Jeggo P. A., and Lobrich M. (2009) ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28, 3413–3427 10.1038/emboj.2009.276 - DOI - PMC - PubMed
-
- Karanam K., Kafri R., Loewer A., and Lahav G. (2012) Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol. Cell 47, 320–329 10.1016/j.molcel.2012.05.052 - V体育平台登录 - DOI - PMC - PubMed
Publication types
- "VSports" Actions
- Actions (VSports手机版)
MeSH terms
- VSports手机版 - Actions
- Actions (V体育ios版)
Grants and funding (VSports手机版)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

