"VSports在线直播" Oncogenic KRAS Regulates Amino Acid Homeostasis and Asparagine Biosynthesis via ATF4 and Alters Sensitivity to L-Asparaginase
- PMID: 29316436
- PMCID: PMC5761662
- DOI: 10.1016/j.ccell.2017.12.003
Oncogenic KRAS Regulates Amino Acid Homeostasis and Asparagine Biosynthesis via ATF4 and Alters Sensitivity to L-Asparaginase
Abstract
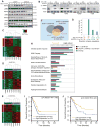
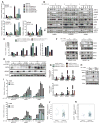
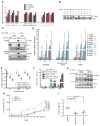
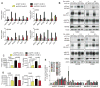
KRAS is a regulator of the nutrient stress response in non-small-cell lung cancer (NSCLC). Induction of the ATF4 pathway during nutrient depletion requires AKT and NRF2 downstream of KRAS VSports手机版. The tumor suppressor KEAP1 strongly influences the outcome of activation of this pathway during nutrient stress; loss of KEAP1 in KRAS mutant cells leads to apoptosis. Through ATF4 regulation, KRAS alters amino acid uptake and asparagine biosynthesis. The ATF4 target asparagine synthetase (ASNS) contributes to apoptotic suppression, protein biosynthesis, and mTORC1 activation. Inhibition of AKT suppressed ASNS expression and, combined with depletion of extracellular asparagine, decreased tumor growth. Therefore, KRAS is important for the cellular response to nutrient stress, and ASNS represents a promising therapeutic target in KRAS mutant NSCLC. .
Keywords: ASNS; ATF4; KEAP1; KRAS; L-asparaginase; NRF2; PI3K; asparagine; glutamine; synthetic vulnerability. V体育安卓版.
Copyright © 2017 Elsevier Inc. All rights reserved. V体育ios版.
"V体育安卓版" Figures



References
-
- Afonyushkin T, Oskolkova OV, Philippova M, Resink TJ, Erne P, Binder BR, Bochkov VN. Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler Thromb Vasc Biol. 2010;30:1007–1013. - PubMed
-
- Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. - PubMed
-
- Arena S, Isella C, Martini M, de Marco A, Medico E, Bardelli A. Knock-in of oncogenic Kras does not transform mouse somatic cells but triggers a transcriptional response that classifies human cancers. Cancer Res. 2007;67:8468–8476. - "VSports注册入口" PubMed
-
- Arnstein HR, Barwick CW, Lange JD, Thomas HD. Control of protein synthesis by amino acid supply. The effect of asparagine deprivation on the translation of messenger RNA in reticulocyte lysates. FEBS letters. 1986;194:146–150. - PubMed
Publication types
- Actions (V体育安卓版)
- Actions (VSports注册入口)
MeSH terms
- Actions (VSports注册入口)
- "VSports最新版本" Actions
- "V体育官网入口" Actions
- "V体育安卓版" Actions
- "VSports手机版" Actions
Substances
Grants and funding
"VSports注册入口" LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

