Secondary Somatic Mutations Restoring RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma
- PMID: 28588062
- PMCID: PMC5612362
- DOI: "VSports手机版" 10.1158/2159-8290.CD-17-0419
Secondary Somatic Mutations Restoring V体育ios版 - RAD51C and RAD51D Associated with Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma
Abstract
High-grade epithelial ovarian carcinomas containing mutated BRCA1 or BRCA2 (BRCA1/2) homologous recombination (HR) genes are sensitive to platinum-based chemotherapy and PARP inhibitors (PARPi), while restoration of HR function due to secondary mutations in BRCA1/2 has been recognized as an important resistance mechanism. We sequenced core HR pathway genes in 12 pairs of pretreatment and postprogression tumor biopsy samples collected from patients in ARIEL2 Part 1, a phase II study of the PARPi rucaparib as treatment for platinum-sensitive, relapsed ovarian carcinoma. In 6 of 12 pretreatment biopsies, a truncation mutation in BRCA1, RAD51C, or RAD51D was identified. In five of six paired postprogression biopsies, one or more secondary mutations restored the open reading frame. Four distinct secondary mutations and spatial heterogeneity were observed for RAD51CIn vitro complementation assays and a patient-derived xenograft, as well as predictive molecular modeling, confirmed that resistance to rucaparib was associated with secondary mutations. Significance: Analyses of primary and secondary mutations in RAD51C and RAD51D provide evidence for these primary mutations in conferring PARPi sensitivity and secondary mutations as a mechanism of acquired PARPi resistance. PARPi resistance due to secondary mutations underpins the need for early delivery of PARPi therapy and for combination strategies. Cancer Discov; 7(9); 984-98. ©2017 AACR. See related commentary by Domchek, p. 937See related article by Quigley et al. , p. 999See related article by Goodall et al. , p. 1006This article is highlighted in the In This Issue feature, p. 920 VSports手机版. .
©2017 American Association for Cancer Research. V体育安卓版.
Conflict of interest statement
Disclosure of Potential Conficts of Interest: A. V. Tinker reports receiving commercial research support from AstraZeneca. M. Friedlander is a consultant/advisory board member for AstraZeneca. D. M. O'Malley is a consultant/advisory board member for Clovis, AstraZeneca, Tesaro, Novocure, Genentech/Roche, Janssen, and Eisai, and is on the steering committee for Amgen. T. C. Harding has ownership interest (including patents) in Clovis Oncology. I. A. McNeish is a consultant/advisory board member for Clovis Oncology. C. L. Scott has received speakers bureau honoraria from Prime Oncology and is a consultant/advisory board member for Clovis Oncology and AstraZeneca. No potential conficts of interest were disclosed by the other authors V体育ios版.
Figures (VSports)

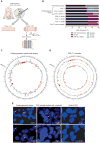
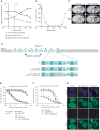
Comment in
-
Reversion Mutations with Clinical Use of PARP Inhibitors: Many Genes, Many Versions.Cancer Discov. 2017 Sep;7(9):937-939. doi: 10.1158/2159-8290.CD-17-0734. Cancer Discov. 2017. PMID: 28864639
References
-
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. - PubMed (V体育官网入口)
-
- Pommier Y, O'Connor MJ, De Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps17. - "VSports" PubMed
-
- Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8. - PMC (V体育平台登录) - PubMed
"VSports手机版" Publication types
- "V体育官网入口" Actions
MeSH terms (V体育ios版)
- "VSports app下载" Actions
- Actions (V体育官网入口)
- "V体育2025版" Actions
- Actions (V体育官网入口)
- Actions (VSports最新版本)
- Actions (V体育ios版)
- "VSports注册入口" Actions
"V体育ios版" Substances
- "V体育安卓版" Actions
- Actions (V体育ios版)
- Actions (VSports最新版本)
- "V体育官网" Actions
VSports - Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
V体育2025版 - Medical
Research Materials
Miscellaneous

