Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1
- PMID: 27928058
- PMCID: PMC5397146
- DOI: 10.1093/nar/gkw1201 (V体育2025版)
Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1
Abstract (V体育安卓版)
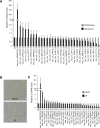
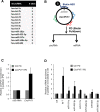
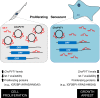
Using RNA sequencing (RNA-Seq), we compared the expression patterns of circular RNAs in proliferating (early-passage) and senescent (late-passage) human diploid WI-38 fibroblasts. Among the differentially expressed senescence-associated circRNAs (which we termed 'SAC-RNAs'), we identified CircPVT1, generated by circularization of an exon of the PVT1 gene, as a circular RNA showing markedly reduced levels in senescent fibroblasts. Reducing CircPVT1 levels in proliferating fibroblasts triggered senescence, as determined by a rise in senescence-associated β-galactosidase activity, higher abundance of CDKN1A/P21 and TP53, and reduced cell proliferation. Although several microRNAs were predicted to bind CircPVT1, only let-7 was found enriched after pulldown of endogenous CircPVT1, suggesting that CircPVT1 might selectively modulate let-7 activity and hence expression of let-7-regulated mRNAs VSports手机版. Reporter analysis revealed that CircPVT1 decreased the cellular pool of available let-7, and antagonizing endogenous let-7 triggered cell proliferation. Importantly, silencing CircPVT1 promoted cell senescence and reversed the proliferative phenotype observed after let-7 function was impaired. Consequently, the levels of several proliferative proteins that prevent senescence, such as IGF2BP1, KRAS and HMGA2, encoded by let-7 target mRNAs, were reduced by silencing CircPVT1. Our findings indicate that the SAC-RNA CircPVT1, elevated in dividing cells and reduced in senescent cells, sequesters let-7 to enable a proliferative phenotype. .
Published by Oxford University Press on behalf of Nucleic Acids Research 2016 V体育安卓版. .
Figures (VSports注册入口)




References
-
- Hayflick L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp. Cell Res. 1965; 37:614–636. - PubMed
-
- Toussaint O., Remacle J., Dierick J.F., Pascal T., Frippiat C., Royer V., Magalhacs J.P., Zdanov S., Chainiaux F.. Stress-induced premature senescence: from biomarkers to likeliness of in vivo occurrence. Biogerontology. 2002; 3:13–17. - PubMed
-
- Busuttil R.A., Dolle M., Campisi J., Vijg J.. Genomic instability, aging, and cellular senescence. Ann. N. Y. Acad. Sci. 2004; 1019:245–255. - "V体育官网" PubMed
-
- Marcotte R., Wang E.. Replicative senescence revisited. J. Gerontol. A Biol. Sci. Med. Sci. 2002; 57:B257–B269. - PubMed
MeSH terms
- V体育ios版 - Actions
- "V体育平台登录" Actions
- VSports手机版 - Actions
- V体育平台登录 - Actions
- Actions (VSports手机版)
Substances
- VSports - Actions
- V体育官网 - Actions
- VSports最新版本 - Actions
- Actions (V体育平台登录)
"V体育官网入口" LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

