The Arkadia-ESRP2 axis suppresses tumor progression: analyses in clear-cell renal cell carcinoma (V体育2025版)
- PMID: 26522722
- PMCID: PMC5399154
- DOI: "VSports" 10.1038/onc.2015.412
"V体育官网" The Arkadia-ESRP2 axis suppresses tumor progression: analyses in clear-cell renal cell carcinoma
Abstract
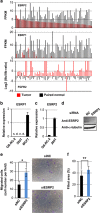
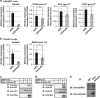
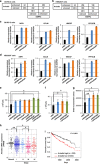
Tumor-specific alternative splicing is implicated in the progression of cancer, including clear-cell renal cell carcinoma (ccRCC). Using ccRCC RNA sequencing data from The Cancer Genome Atlas, we found that epithelial splicing regulatory protein 2 (ESRP2), one of the key regulators of alternative splicing in epithelial cells, is expressed in ccRCC. ESRP2 mRNA expression did not correlate with the overall survival rate of ccRCC patients, but the expression of some ESRP-target exons correlated with the good prognosis and with the expression of Arkadia (also known as RNF111) in ccRCC. Arkadia physically interacted with ESRP2, induced polyubiquitination and modulated its splicing function VSports手机版. Arkadia and ESRP2 suppressed ccRCC tumor growth in a coordinated manner. Lower expression of Arkadia correlated with advanced tumor stages and poor outcomes in ccRCC patients. This study thus reveals a novel tumor-suppressive role of the Arkadia-ESRP2 axis in ccRCC. .
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gamazon ER, Stranger BE. Genomics of alternative splicing: evolution, development and pathophysiology. Hum Genet 2014; 133: 679–687. - PubMed
-
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 2011; 12: 715–729. - "V体育平台登录" PMC - PubMed
-
- Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene 2014; 34: 1–14. - PubMed
-
- Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Semin Cell Dev Biol 2014; 32C: 30–36. - PubMed
-
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010; 10: 116–129. - PubMed
Publication types (VSports最新版本)
- Actions (VSports app下载)
MeSH terms
- "V体育官网入口" Actions
- "V体育平台登录" Actions
- VSports - Actions
- Actions (VSports最新版本)
- Actions (VSports)
- Actions (V体育官网)
- Actions (VSports在线直播)
- V体育ios版 - Actions
- "V体育官网" Actions
Substances
- "VSports在线直播" Actions
- "V体育官网" Actions
- "V体育官网入口" Actions
LinkOut - more resources
Full Text Sources (V体育官网)
Other Literature Sources (VSports)
Medical
Molecular Biology Databases
VSports - Research Materials

