Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase (V体育官网入口)
- PMID: 26365377
- PMCID: PMC4575890
- DOI: 10.1016/j.molcel.2015.07.029
Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase (VSports)
Abstract (V体育ios版)
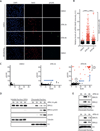
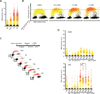
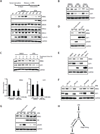
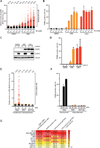
The ATR-Chk1 pathway is critical for DNA damage responses and cell-cycle progression. Chk1 inhibition is more deleterious to cycling cells than ATR inhibition, raising questions about ATR and Chk1 functions in the absence of extrinsic replication stress. Here we show that a key role of ATR in S phase is to coordinate RRM2 accumulation and origin firing VSports手机版. ATR inhibitor (ATRi) induces massive ssDNA accumulation and replication catastrophe in a fraction of early S-phase cells. In other S-phase cells, however, ATRi induces moderate ssDNA and triggers a DNA-PK and Chk1-mediated backup pathway to suppress origin firing. The backup pathway creates a threshold such that ATRi selectively kills cells under high replication stress, whereas Chk1 inhibitor induces cell death at a lower threshold. The levels of ATRi-induced ssDNA correlate with ATRi sensitivity in a panel of cell lines, suggesting that ATRi-induced ssDNA could be predictive of ATRi sensitivity in cancer cells. .
Copyright © 2015 Elsevier Inc V体育安卓版. All rights reserved. .
Figures



References
-
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. - PubMed
-
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. - PubMed
-
- Bastos de Oliveira FM, Kim D, Cussiol JR, Das J, Jeong MC, Doerfler L, Schmidt KH, Yu H, Smolka MB. Phosphoproteomics Reveals Distinct Modes of Mec1/ATR Signaling during DNA Replication. Mol Cell. 2015;57:1124–1132. - PMC (VSports最新版本) - PubMed
-
- Beck H, Nahse-Kumpf V, Larsen MS, O’Hanlon KA, Patzke S, Holmberg C, Mejlvang J, Groth A, Nielsen O, Syljuasen RG, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–4236. - VSports - PMC - PubMed
Publication types
MeSH terms
- VSports最新版本 - Actions
- Actions (VSports注册入口)
- "VSports" Actions
- "VSports最新版本" Actions
- "VSports" Actions
- "V体育安卓版" Actions
- Actions (V体育官网入口)
- "VSports最新版本" Actions
Substances
- "VSports手机版" Actions
- "VSports最新版本" Actions
- "VSports在线直播" Actions
- "V体育平台登录" Actions
"VSports" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
"V体育ios版" Miscellaneous

