Emergent Self-Organized Criticality in Gene Expression Dynamics: Temporal Development of Global Phase Transition Revealed in a Cancer Cell Line
- PMID: 26067993
- PMCID: PMC4465832 (VSports app下载)
- DOI: V体育安卓版 - 10.1371/journal.pone.0128565
Emergent Self-Organized Criticality in Gene Expression Dynamics: Temporal Development of Global Phase Transition Revealed in a Cancer Cell Line
Abstract
Background: The underlying mechanism of dynamic control of the genome-wide expression is a fundamental issue in bioscience. We addressed it in terms of phase transition by a systemic approach based on both density analysis and characteristics of temporal fluctuation for the time-course mRNA expression in differentiating MCF-7 breast cancer cells. VSports手机版.
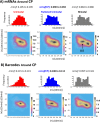
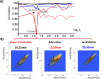
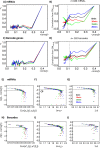
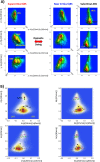
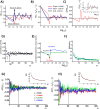
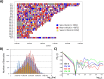
Methodology: In a recent work, we suggested criticality as an essential aspect of dynamic control of genome-wide gene expression. Criticality was evident by a unimodal-bimodal transition through flattened unimodal expression profile. The flatness on the transition suggests the existence of a critical transition at which up- and down-regulated expression is balanced. Mean field (averaging) behavior of mRNAs based on the temporal expression changes reveals a sandpile type of transition in the flattened profile V体育安卓版. Furthermore, around the transition, a self-similar unimodal-bimodal transition of the whole expression occurs in the density profile of an ensemble of mRNA expression. These singular and scaling behaviors identify the transition as the expression phase transition driven by self-organized criticality (SOC). .
Principal findings: Emergent properties of SOC through a mean field approach are revealed: i) SOC, as a form of genomic phase transition, consolidates distinct critical states of expression, ii) Coupling of coherent stochastic oscillations between critical states on different time-scales gives rise to SOC, and iii) Specific gene clusters (barcode genes) ranging in size from kbp to Mbp reveal similar SOC to genome-wide mRNA expression and ON-OFF synchronization to critical states. This suggests that the cooperative gene regulation of topological genome sub-units is mediated by the coherent phase transitions of megadomain-scaled conformations between compact and swollen chromatin states V体育ios版. .
Conclusion and significance: In summary, our study provides not only a systemic method to demonstrate SOC in whole-genome expression, but also introduces novel, physically grounded concepts for a breakthrough in the study of biological regulation. VSports最新版本.
Conflict of interest statement (V体育ios版)
Figures




References
-
- Klevecz RR, Bolen J, Forrest G, Murray DB (2004) A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA. 100: 1200–1205. - PMC (VSports) - PubMed
-
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 131: 861–872. - PubMed
-
- Young RA (2011) Control of the embryonic stem cell state. Cell 144: 940–945. 10.1016/j.cell.2011.01.032 - DOI (VSports) - PMC - PubMed
Publication types (VSports手机版)
MeSH terms
- VSports app下载 - Actions
- Actions (VSports手机版)
- "V体育官网" Actions
- V体育平台登录 - Actions
- V体育官网入口 - Actions
- VSports最新版本 - Actions
- V体育平台登录 - Actions
Substances
- Actions (V体育平台登录)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

