A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines
- PMID: 25960936
- PMCID: PMC4355981
- DOI: 10.4161/21624011.2014.954893
A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines
Abstract
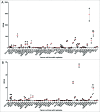
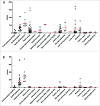
Cancer cell lines are a tremendous resource for cancer biology and therapy development. These multipurpose tools are commonly used to examine the genetic origin of cancers, to identify potential novel tumor targets, such as tumor antigens for vaccine devel-opment, and utilized to screen potential therapies in preclinical studies. Mutations, gene expression, and drug sensitivity have been determined for many cell lines using next-generation sequencing (NGS). However, the human leukocyte antigen (HLA) type and HLA expression of tumor cell lines, characterizations necessary for the development of cancer vaccines, have remained largely incomplete and, such information, when available, has been distributed in many publications. Here, we determine the 4-digit HLA type and HLA expression of 167 cancer and 10 non-cancer cell lines from publically available RNA-Seq data. We use standard NGS RNA-Seq short reads from "whole transcriptome" sequencing, map reads to known HLA types, and statistically determine HLA type, heterozygosity, and expression. First, we present previously unreported HLA Class I and II genotypes. Second, we determine HLA expression levels in each cancer cell line, providing insights into HLA downregulation and loss in cancer. Third, using these results, we provide a fundamental cell line "barcode" to track samples and prevent sample annotation swaps and contamination. Fourth, we integrate the cancer cell-line specific HLA types and HLA expression with available cell-line specific mutation information and existing HLA binding prediction algorithms to make a catalog of predicted antigenic mutations in each cell line. The compilation of our results are a fundamental resource for all researchers selecting specific cancer cell lines based on the HLA type and HLA expression, as well as for the development of immunotherapeutic tools for novel cancer treatment modalities VSports手机版. .
Keywords: BRENDA, BRaunschweig ENzyme Database; CCLE, Cancer Cell Line Encyclopedia; COSMIC, Catalog of Somatic Mutations in Cancer; DLBCL, diffuse large B-cell lymphoma; HLA expression; HLA type; HLA, Human Leukocyte Antigen; IEDB, Immune Epitope Database; NGS, Next Generation Sequencing; RNA-Seq; RNA-Seq, RNA Sequencing; RPKM, reads per kilobase of exon model per million mapped reads; SNV, single nucleotide variation; SRA, Sequence Read Archive; cancer cell lines; immunotherapy; neoepitopes; nsSNV, non synonymous SNV; somatic mutations. V体育安卓版.
Figures


References
-
- Gillet J.-P., Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst 2013; 105:452-8; PMID:; http://dx.doi.org/10.1093/jnci/djt007 - DOI - PMC - PubMed
-
- Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer 2010; 10:241-53; PMID:; http://dx.doi.org/10.1038/nrc2820 - DOI - PubMed
-
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. . COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011; 39:D945-50; PMID:; "V体育2025版" http://dx.doi.org/10.1093/nar/gkq929 - DOI - PMC - PubMed
-
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. . The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483:603-7; PMID:; VSports最新版本 - http://dx.doi.org/10.1038/nature11003 - DOI - PMC - PubMed
-
- Castle JC, Loewer M, Boegel S, de Graaf J, Bender C, Tadmor, Boisguerin V, Bukur T, Sorn P, Paret C, et al. . Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics 2014; 15:190; PMID:; "V体育安卓版" http://dx.doi.org/10.1186/1471-2164-15-190 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
V体育官网入口 - Full Text Sources
Other Literature Sources
VSports注册入口 - Research Materials
