Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway
- PMID: 21816818
- PMCID: PMC3190819
- DOI: 10.1074/jbc.M111.274159
Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway
Abstract
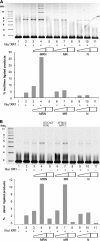
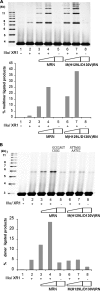
Recent studies have implicated a poorly defined alternative pathway of nonhomologous end joining (alt-NHEJ) in the generation of large deletions and chromosomal translocations that are frequently observed in cancer cells. Here, we describe an interaction between two factors, hMre11/hRad50/Nbs1 (MRN) and DNA ligase IIIα/XRCC1, that have been linked with alt-NHEJ. Expression of DNA ligase IIIα and the association between MRN and DNA ligase IIIα/XRCC1 are altered in cell lines defective in the major NHEJ pathway. Most notably, DNA damage induced the association of these factors in DNA ligase IV-deficient cells. MRN interacts with DNA ligase IIIα/XRCC1, stimulating intermolecular ligation, and together these proteins join incompatible DNA ends in a reaction that mimics alt-NHEJ. Thus, our results provide novel mechanistic insights into the alt-NHEJ pathway that not only contributes to genome instability in cancer cells but may also be a therapeutic target VSports手机版. .
Figures (VSports注册入口)





V体育平台登录 - References
-
- Venkitaraman A. R. (2002) Cell 108, 171–182 - PubMed
-
- Sharpless N. E., Ferguson D. O., O'Hagan R. C., Castrillon D. H., Lee C., Farazi P. A., Alson S., Fleming J., Morton C. C., Frank K., Chin L., Alt F. W., DePinho R. A. (2001) Mol. Cell 8, 1187–1196 - PubMed
-
- Lieber M. R. (2008) J. Biol. Chem. 283, 1–5 - PubMed
-
- Weterings E., Chen D. J. (2008) Cell Res. 18, 114–124 - PubMed
"VSports注册入口" Publication types
- V体育官网入口 - Actions
- "VSports手机版" Actions
"V体育平台登录" MeSH terms
- VSports在线直播 - Actions
- Actions (VSports在线直播)
- "VSports在线直播" Actions
- Actions (V体育平台登录)
- VSports手机版 - Actions
- "V体育官网" Actions
- "V体育官网入口" Actions
- VSports最新版本 - Actions
Substances
- Actions (V体育平台登录)
- V体育2025版 - Actions
- VSports app下载 - Actions
- "VSports手机版" Actions
- Actions (V体育安卓版)
- "VSports手机版" Actions
- Actions (V体育官网入口)
- V体育平台登录 - Actions
Grants and funding
VSports在线直播 - LinkOut - more resources
Full Text Sources
Molecular Biology Databases (VSports最新版本)
Research Materials
Miscellaneous

