Palmitoylation of ketogenic enzyme HMGCS2 enhances its interaction with PPARalpha and transcription at the Hmgcs2 PPRE
- PMID: 20124434
- PMCID: PMC2874477
- DOI: 10.1096/fj.09-149765
Palmitoylation of ketogenic enzyme HMGCS2 enhances its interaction with PPARalpha and transcription at the Hmgcs2 PPRE
Abstract
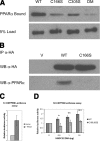
Excessive liver production of ketone bodies is one of many metabolic complications that can arise from diabetes, and in severe untreated cases, it can result in ketoacidosis, coma, and death. Mitochondrial HMG-CoA synthase (HMGCS2), the rate-limiting enzyme in ketogenesis, has been shown to interact with PPARalpha and act as a coactivator to up-regulate transcription from the PPRE of its own gene. Although protein palmitoylation is typically a cytosolic process that promotes membrane association, we recently identified 21 palmitoylated proteins in rat liver mitochondria, including HMGCS2 VSports手机版. Herein, our data support a mechanism whereby palmitate is first added onto HMGCS2 active site Cys166 and then transacylated to Cys305. Palmitoylation promotes the HMGCS2/PPARalpha interaction, resulting in transcriptional activation from the Hmgcs2 PPRE. These results, together with the fact that 8 of the 21 palmitoylated mitochondrial proteins that we previously identified have nuclear receptor interacting motifs, demonstrate a novel--and perhaps ubiquitous--role for palmitoylation as a modulator of transcription. .
Figures




References
-
- Cullingford T E. The ketogenic diet; fatty acids, fatty acid-activated receptors and neurological disorders. Prostaglandins Leukot Essent Fatty Acids. 2004;70:253–264. - PubMed
-
- Fukao T, Lopaschuk G D, Mitchell G A. Pathways and control of ketone body metabolism: on the fringe of lipid biochemistry. Prostaglandins Leukot Essent Fatty Acids. 2004;70:243–251. - VSports - PubMed
-
- Eledrisi M S, Alshanti M S, Shah M F, Brolosy B, Jaha N. Overview of the diagnosis and management of diabetic ketoacidosis. Am J Med Sci. 2006;331:243–251. - PubMed
-
- Wolfsdorf J, Craig M E, Daneman D, Dunger D, Edge J, Lee W R, Rosenbloom A, Sperling M A, Hanas R. Diabetic ketoacidosis. Pediatr Diabetes. 2007;8:28–43. - PubMed
Publication types
MeSH terms
- VSports注册入口 - Actions
- Actions (VSports最新版本)
- "VSports在线直播" Actions
- V体育平台登录 - Actions
- "V体育ios版" Actions
- Actions (V体育平台登录)
- Actions (VSports手机版)
- Actions (VSports app下载)
- VSports - Actions
- "V体育官网" Actions
Substances
- "V体育ios版" Actions
- VSports手机版 - Actions
- "V体育官网入口" Actions
Grants and funding (V体育官网)
LinkOut - more resources
"VSports最新版本" Full Text Sources

