V体育官网入口 - MicroRNA-34a is an important component of PRIMA-1-induced apoptotic network in human lung cancer cells
- PMID: 19921694
- PMCID: VSports注册入口 - PMC4153870
- DOI: 10.1002/ijc.25049
"VSports最新版本" MicroRNA-34a is an important component of PRIMA-1-induced apoptotic network in human lung cancer cells
Abstract
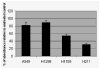
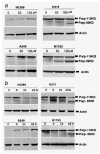
Restoration of p53 function in tumor cells would be an attractive strategy for lung cancer therapy because p53 mutations are found in more than 50% of lung cancers. The small molecule PRIMA-1 has been shown to restore the tumor suppression function of p53 and to induce apoptosis in human tumor cells. The mechanism of apoptosis induced by PRIMA-1 remains unclear. We investigated the effects of PRIMA-1 in apoptosis with Western immunoblot analysis, TaqMan microRNA real-time PCR, cell viability analysis and flow cytometry using human lung cancer cell lines containing mutant (H211 and H1155), wild-type (A549) or null (H1299) p53. PRIMA-1 induced massive apoptosis in the H211 and H1155 cells, but was less toxic to the A549 and H1299 cells VSports手机版. Western immunoblot analysis showed cleavage of PARP in H211 and H1155 cells but not in A549 and H1299 cells following treatment with PRIMA-1. In addition, p53 protein was also phosphorylated in H211 and H1155 cells. TaqMan microRNA assay showed that the expression of microRNA-34a was increased in the H211 and H1155 cells posttreatment. Knockdown microRNA-34a decreased the rate of apoptosis caused by PRIMA-1. The above results suggest that microRNA-34a is one of the important components of PRIMA-1-induced apoptotic network in the cancer cells harboring mutant p53. .
Figures



References
-
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. - "VSports在线直播" PubMed
-
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. - PubMed
-
- El-Hizawi S, Lagowski JP, Kulesz-Martin M, Albor A. Induction of gene amplification as a gain-of-function phenotype of mutant p53 proteins. Cancer Res. 2002;62:3264–70. - V体育2025版 - PubMed
-
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–14. - "VSports app下载" PubMed
-
- Mitsudomi T, Steinberg SM, Nau MM, Carbone D, D’Amico D, Bodner S, Oie HK, Linnoila RI, Mulshine JL, Minna JD, Gazdar AF. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–80. - VSports在线直播 - PubMed
VSports最新版本 - Publication types
- "VSports最新版本" Actions
MeSH terms (VSports注册入口)
- "V体育官网入口" Actions
- Actions (VSports在线直播)
- Actions (VSports注册入口)
- "VSports最新版本" Actions
- "VSports最新版本" Actions
- V体育官网 - Actions
- "VSports" Actions
Substances
- V体育平台登录 - Actions
- Actions (VSports最新版本)
- V体育官网 - Actions
- V体育ios版 - Actions
- "VSports app下载" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
V体育平台登录 - Research Materials
Miscellaneous

