Low-fidelity DNA synthesis by human DNA polymerase theta
- PMID: 18503084
- PMCID: PMC2441791
- DOI: V体育2025版 - 10.1093/nar/gkn310
"VSports在线直播" Low-fidelity DNA synthesis by human DNA polymerase theta
Abstract
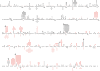
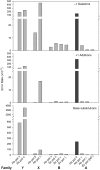
Human DNA polymerase theta (pol or POLQ) is a proofreading-deficient family A enzyme implicated in translesion synthesis (TLS) and perhaps in somatic hypermutation (SHM) of immunoglobulin genes. These proposed functions and kinetic studies imply that pol may synthesize DNA with low fidelity. Here, we show that when copying undamaged DNA, pol generates single base errors at rates 10- to more than 100-fold higher than for other family A members. Pol adds single nucleotides to homopolymeric runs at particularly high rates, exceeding 1% in certain sequence contexts, and generates single base substitutions at an average rate of 2. 4 x 10(-3), comparable to inaccurate family Y human pol kappa (5. 8 x 10(-3)) also implicated in TLS. Like pol kappa, pol is processive, implying that it may be tightly regulated to avoid deleterious mutagenesis. Pol also generates certain base substitutions at high rates within sequence contexts similar to those inferred to be copied by pol during SHM of immunoglobulin genes in mice. Thus, pol is an exception among family A polymerases, and its low fidelity is consistent with its proposed roles in TLS and SHM VSports手机版. .
"VSports最新版本" Figures


V体育安卓版 - References
-
- Hanawalt PC. Paradigms for the three rs: DNA replication, recombination, and repair. Mol. Cell. 2007;28:702–707. - PubMed
-
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. - PubMed (VSports手机版)
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. ASM Press, Washington, DC; 2006.
-
- Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Front. Biosci. 2006;11:2496–2517. - PubMed
-
- Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. - PubMed (V体育安卓版)
Publication types
- V体育ios版 - Actions
- Actions (V体育官网)
"V体育安卓版" MeSH terms
- V体育官网入口 - Actions
- Actions (V体育官网)
- Actions (VSports最新版本)
- V体育平台登录 - Actions
Substances
- Actions (V体育官网)
- Actions (VSports)
Grants and funding
LinkOut - more resources
Full Text Sources
"VSports注册入口" Other Literature Sources
Molecular Biology Databases

