Improved breast cancer prognosis through the combination of clinical and genetic markers
- PMID: 17130137
- PMCID: PMC3431620
- DOI: "VSports最新版本" 10.1093/bioinformatics/btl543
Improved breast cancer prognosis through the combination of clinical and genetic markers
Abstract
Motivation: Accurate prognosis of breast cancer can spare a significant number of breast cancer patients from receiving unnecessary adjuvant systemic treatment and its related expensive medical costs. Recent studies have demonstrated the potential value of gene expression signatures in assessing the risk of post-surgical disease recurrence. However, these studies all attempt to develop genetic marker-based prognostic systems to replace the existing clinical criteria, while ignoring the rich information contained in established clinical markers. Given the complexity of breast cancer prognosis, a more practical strategy would be to utilize both clinical and genetic marker information that may be complementary VSports手机版. .
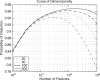
Methods: A computational study is performed on publicly available microarray data, which has spawned a 70-gene prognostic signature. The recently proposed I-RELIEF algorithm is used to identify a hybrid signature through the combination of both genetic and clinical markers V体育安卓版. A rigorous experimental protocol is used to estimate the prognostic performance of the hybrid signature and other prognostic approaches. Survival data analyses is performed to compare different prognostic approaches. .
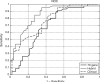
Results: The hybrid signature performs significantly better than other methods, including the 70-gene signature, clinical makers alone and the St. Gallen consensus criterion. At the 90% sensitivity level, the hybrid signature achieves 67% specificity, as compared to 47% for the 70-gene signature and 48% for the clinical makers V体育ios版. The odds ratio of the hybrid signature for developing distant metastases within five years between the patients with a good prognosis signature and the patients with a bad prognosis is 21. 0 (95% CI:6. 5-68. 3), far higher than either genetic or clinical markers alone. .
Availability: The breast cancer dataset is available at www. nature. com and Matlab codes are available upon request. VSports最新版本.
Figures

References
-
- Abba M, et al. Gene expression signature of estrogen receptor α status in breast cancer. BMC Genomics. 2005;6:74–81. - V体育2025版 - PMC - PubMed
-
- Brenton J, et al. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J. Clin. Oncol. 2005;23:7350–7360. - "VSports app下载" PubMed
-
- Dalton W, et al. Cancer biomarkers–an invitation to the table. Science. 2006;312:1165–1168. - PubMed
-
- de Mascarel I, et al. Obvious peritumorous emboli: an elusive prognostic factor reappraised: multivariate analysis of 1320 node-negative breast cancers. Eur. J. Cancer. 1998;34:58–65. - PubMed
-
- Dettling M, et al. Finding predictive gene groups from microarray data. J. Multivariate Anal. 2004;1:106–131.
MeSH terms
- "V体育2025版" Actions
- "VSports手机版" Actions
- Actions (V体育平台登录)
- Actions (VSports在线直播)
- Actions (VSports最新版本)
- Actions (V体育官网入口)
- "VSports手机版" Actions
- Actions (VSports注册入口)
- Actions (VSports手机版)
Substances
Grants and funding
LinkOut - more resources (V体育官网入口)
"VSports最新版本" Full Text Sources
Other Literature Sources
Medical (VSports最新版本)
Research Materials

