Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes
- PMID: 15060169
- PMCID: VSports最新版本 - PMC381674
- DOI: "VSports最新版本" 10.1128/MCB.24.8.3505-3513.2004
Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes
Abstract
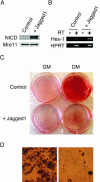
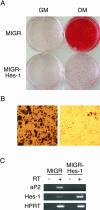
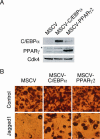
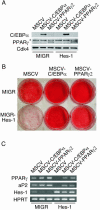
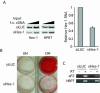
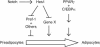
The process of adipogenesis involves a complex program of gene expression that includes down-regulation of the gene encoding Hes-1, a target of the Notch signaling pathway. To determine if Notch signaling affects adipogenesis, we exposed 3T3-L1 preadipocytes to the Notch ligand Jagged1 and found that differentiation was significantly reduced. This effect could be mimicked by constitutive expression of Hes-1 VSports手机版. The block was associated with a complete loss of C/EBPalpha and peroxisome proliferator-activated receptor gamma (PPARgamma) induction and could be overcome by retroviral expression of either C/EBPalpha or PPARgamma2. Surprisingly, small interfering RNA (siRNA)-mediated reduction of Hes-1 mRNA in 3T3-L1 cells also inhibited differentiation, suggesting an additional, obligatory role for Hes-1 in adipogenesis. This role may be related to our observation that both Notch signaling and Hes-1 down-regulate transcription of the gene encoding DLK/Pref-1, a protein known to inhibit differentiation of 3T3-L1 cells. The results presented in this study establish a new target downstream of the Notch-Hes-1 pathway and suggest a dual role for Hes-1 in adipocyte development. .
Figures


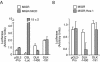
References
-
- Aster, J. C., E. S. Robertson, R. P. Hasserjian, J. R. Turner, E. Kieff, and J. Sklar. 1997. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J. Biol. Chem. 272:11336-11343. - PubMed
-
- Berezovska, O., C. Jack, P. McLean, J. C. Aster, C. Hicks, W. Xia, M. S. Wolfe, W. T. Kimberly, G. Weinmaster, D. J. Selkoe, and B. T. Hyman. 2000. Aspartate mutations in presenilin and gamma-secretase inhibitors both impair notch1 proteolysis and nuclear translocation with relative preservation of notch1 signaling. J. Neurochem. 75:583-593. - PubMed
-
- Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5:207-216. - V体育官网入口 - PubMed
-
- De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518-522. - PubMed
"V体育安卓版" Publication types
MeSH terms
- "VSports app下载" Actions
- "V体育官网" Actions
- V体育安卓版 - Actions
- "V体育2025版" Actions
- "VSports" Actions
- VSports app下载 - Actions
- Actions (VSports最新版本)
- Actions (V体育平台登录)
- Actions (VSports)
- Actions (VSports)
- VSports手机版 - Actions
- VSports注册入口 - Actions
- "V体育安卓版" Actions
- "VSports最新版本" Actions
- "V体育官网入口" Actions
- Actions (V体育平台登录)
Substances
- Actions (VSports手机版)
- VSports最新版本 - Actions
- Actions (V体育平台登录)
- V体育ios版 - Actions
- Actions (V体育安卓版)
- V体育官网 - Actions
- "V体育安卓版" Actions
- "VSports" Actions
- "V体育2025版" Actions
"VSports在线直播" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
