Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication
- PMID: 9990856
- PMCID: "V体育ios版" PMC316435
- DOI: 10.1101/gad.13.3.320
"V体育官网入口" Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication
"VSports最新版本" Abstract
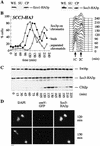
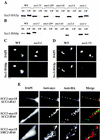
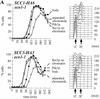
Sister chromatid cohesion is crucial for chromosome segregation during mitosis. Loss of cohesion very possibly triggers sister separation at the metaphase --> anaphase transition. This process depends on the destruction of anaphase inhibitory proteins like Pds1p (Cut2p), which is thought to liberate a sister-separating protein Esp1p (Cut1p). By looking for mutants that separate sister centromeres in the presence of Pds1p, this and a previous study have identified six proteins essential for establishing or maintaining sister chromatid cohesion VSports手机版. Four of these proteins, Scc1p, Scc3p, Smc1p, and Smc3p, are subunits of a 'Cohesin' complex that binds chromosomes from late G1 until the onset of anaphase. The fifth protein, Scc2p, is not a stoichiometric Cohesin subunit but it is required for Cohesin's association with chromosomes. The sixth protein, Eco1p(Ctf7p), is not a Cohesin subunit. It is necessary for the establishment of cohesion during DNA replication but not for its maintenance during G2 and M phases. .
Figures









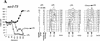
VSports app下载 - References
-
- Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmids: A uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. - PubMed
-
- Birkenbihl RP, Subramani S. The rad21 gene product of Schizosacharomyces pombe is a nuclear, cell cycle regulated phosphoprotein. J Cell Biol. 1995;270:7703–7711. - PubMed
-
- Carramolino L, Lee B, Zaballos A, Peled A, Barthelemy I, Shav-Tal Y, Prieto I, Carmi P, Gothelf Y, Gonzalez de Buitrago G, Aracil M, Marquez G, Barbero J, D Z. SA-1, a nuclear protein encoded by one member of a novel gene family: Molecular cloning and detection in hemopoietic organs. Gene. 1997;195:151–159. - "V体育官网" PubMed
-
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An Esp1/Pds1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. - PubMed
-
- Cohen-Fix O, Peters J-M, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. - "VSports注册入口" PubMed
"VSports在线直播" Publication types
MeSH terms (VSports)
- Actions (VSports)
- Actions (V体育ios版)
- Actions (VSports注册入口)
- "V体育2025版" Actions
- "VSports手机版" Actions
- VSports app下载 - Actions
- "V体育平台登录" Actions
- VSports手机版 - Actions
- "VSports" Actions
- "V体育官网入口" Actions
- V体育官网入口 - Actions
Substances
- V体育官网 - Actions
- Actions (VSports在线直播)
- "V体育平台登录" Actions
- VSports最新版本 - Actions
LinkOut - more resources
VSports最新版本 - Full Text Sources
Molecular Biology Databases (V体育ios版)
