Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo
- PMID: 9786943
- PMCID: "VSports" PMC2132840
- DOI: V体育官网 - 10.1083/jcb.143.2.297
Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo (VSports注册入口)
Abstract
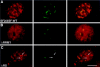
Expression of most RNA polymerase II transcripts requires the coordinated execution of transcription, splicing, and 3' processing. We have previously shown that upon transcriptional activation of a gene in vivo, pre-mRNA splicing factors are recruited from nuclear speckles, in which they are concentrated, to sites of transcription (Misteli, T. , J. F. Cáceres, and D. L VSports手机版. Spector. 1997. Nature. 387:523-527). This recruitment process appears to spatially coordinate transcription and pre-mRNA splicing within the cell nucleus. Here we have investigated the molecular basis for recruitment by analyzing the recruitment properties of mutant splicing factors. We show that multiple protein domains are required for efficient recruitment of SR proteins from nuclear speckles to nascent RNA. The two types of modular domains found in the splicing factor SF2/ ASF exert distinct functions in this process. In living cells, the RS domain functions in the dissociation of the protein from speckles, and phosphorylation of serine residues in the RS domain is a prerequisite for this event. The RNA binding domains play a role in the association of splicing factors with the target RNA. These observations identify a novel in vivo role for the RS domain of SR proteins and suggest a model in which protein phosphorylation is instrumental for the recruitment of these proteins to active sites of transcription in vivo. .
Figures



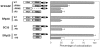

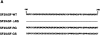
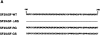
References
-
- Báuren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. - "VSports注册入口" PubMed
-
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. - PubMed
V体育安卓版 - Publication types
- VSports在线直播 - Actions
MeSH terms
- "V体育平台登录" Actions
- "VSports在线直播" Actions
- V体育ios版 - Actions
- V体育官网 - Actions
- Actions (V体育平台登录)
- "V体育平台登录" Actions
- "V体育2025版" Actions
- "V体育平台登录" Actions
- VSports手机版 - Actions
- VSports最新版本 - Actions
Substances
- Actions (VSports在线直播)
- V体育平台登录 - Actions
Grants and funding
LinkOut - more resources
VSports app下载 - Full Text Sources
Other Literature Sources
"VSports注册入口" Research Materials

