"V体育官网" Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes
- PMID: 9362057
- PMCID: PMC25696 (V体育安卓版)
- DOI: 10.1091/mbc.8.11.2119
Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes
Abstract
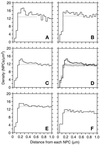
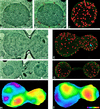
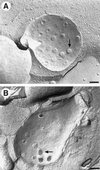
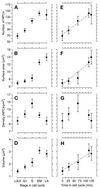
The number of nuclear pore complexes (NPCs) in individual nuclei of the yeast Saccharomyces cerevisiae was determined by computer-aided reconstruction of entire nuclei from electron micrographs of serially sectioned cells. Nuclei of 32 haploid cells at various points in the cell cycle were modeled and found to contain between 65 and 182 NPCs. Morphological markers, such as cell shape and nuclear shape, were used to determine the cell cycle stage of the cell being examined VSports手机版. NPC number was correlated with cell cycle stage to reveal that the number of NPCs increases steadily, beginning in G1-phase, suggesting that NPC assembly occurs continuously throughout the cell cycle. However, accumulation of nuclear envelope observed during the cell cycle, indicated by nuclear surface area, is not continuous at the same rate, such that the density of NPCs per unit area of nuclear envelope peaks in apparent S-phase cells. Analysis of the nuclear envelope reconstructions also revealed no preferred NPC-to-NPC distance. However, NPCs were found in large clusters over regions of the nuclear envelope. Interestingly, clusters of NPCs were most pronounced in early mitotic nuclei and were found to be associated with the spindle pole bodies, but the functional significance of this association is unknown. .
V体育安卓版 - Figures


References
-
- Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. Molecular Biology of the Yeast Saccharomyces. I. Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 59–96.
-
- Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. - PubMed
Publication types
VSports注册入口 - MeSH terms
- Actions (VSports最新版本)
- "VSports最新版本" Actions
V体育安卓版 - Grants and funding
LinkOut - more resources
V体育ios版 - Full Text Sources
"V体育ios版" Molecular Biology Databases

