"VSports最新版本" Model Cell Lines and Tissues of Different HGSOC Subtypes Differ in Local Estrogen Biosynthesis
- PMID: 35681563
- PMCID: PMC9179372
- DOI: 10.3390/cancers14112583
Model Cell Lines and Tissues of Different HGSOC Subtypes Differ in Local Estrogen Biosynthesis
Abstract (V体育2025版)
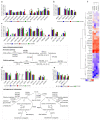
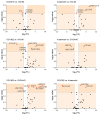
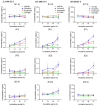
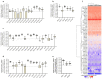
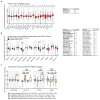
Ovarian cancer (OC) is highly lethal and heterogeneous. Several hormones are involved in OC etiology including estrogens; however, their role in OC is not completely understood. Here, we performed targeted transcriptomics and estrogen metabolism analyses in high-grade serous OC (HGSOC), OVSAHO, Kuramochi, COV632, and immortalized normal ovarian epithelial HIO-80 cells. We compared these data with public transcriptome and proteome data for the HGSOC tissues. In all model systems, high steroid sulfatase expression and weak/undetected aromatase (CYP19A1) expression indicated the formation of estrogens from the precursor estrone-sulfate (E1-S). In OC cells, the metabolism of E1-S to estradiol was the highest in OVSAHO, followed by Kuramochi and COV362 cells, and decreased with increasing chemoresistance. In addition, higher HSD17B14 and CYP1A2 expressions were observed in highly chemoresistant COV362 cells and platinum-resistant tissues compared to those in HIO-80 cells and platinum-sensitive tissues. The HGSOC cell models differed in HSD17B10, CYP1B1, and NQO1 expression. Proteomic data also showed different levels of HSD17B10, CYP1B1, NQO1, and SULT1E1 between the four HGSOC subtypes. These results suggest that different HGSOC subtypes form different levels of estrogens and their metabolites and that the estrogen-biosynthesis-associated targets should be further studied for the development of personalized treatment VSports手机版. .
Keywords: COV362; HIO-80; Kuramochi; OVSAHO; differentiated; high-grade serous ovarian carcinoma; immunoreactive; mesenchymal subtype; ovarian cancer; proliferative. V体育安卓版.
Conflict of interest statement
The authors declare no conflict of interest.
Figures (VSports最新版本)

References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. - "VSports" DOI - PubMed
-
- Desai A., Xu J., Aysola K., Qin Y., Okoli C., Hariprasad R., Chinemerem U., Gates C., Reddy A., Danner O., et al. Epithelial ovarian cancer: An overview. World J. Transl. Med. 2014;3:1–8. doi: 10.5528/wjtm.v3.i1.1. - DOI (VSports最新版本) - PMC - PubMed
Grants and funding (VSports在线直播)
LinkOut - more resources
Full Text Sources
"VSports在线直播" Research Materials
Miscellaneous

