"VSports最新版本" NEAT-seq: simultaneous profiling of intra-nuclear proteins, chromatin accessibility and gene expression in single cells
- PMID: 35501385
- PMCID: PMC11192021 (V体育平台登录)
- DOI: 10.1038/s41592-022-01461-y
NEAT-seq: simultaneous profiling of intra-nuclear proteins, chromatin accessibility and gene expression in single cells
Abstract
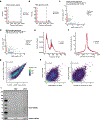
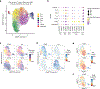
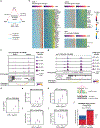
In this work, we describe NEAT-seq (sequencing of nuclear protein epitope abundance, chromatin accessibility and the transcriptome in single cells), enabling interrogation of regulatory mechanisms spanning the central dogma VSports手机版. We apply this technique to profile CD4 memory T cells using a panel of master transcription factors (TFs) that drive T cell subsets and identify examples of TFs with regulatory activity gated by transcription, translation and regulation of chromatin binding. We also link a noncoding genome-wide association study single-nucleotide polymorphism (SNP) within a GATA motif to a putative target gene, using NEAT-seq data to internally validate SNP impact on GATA3 regulation. .
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc. V体育安卓版.
Conflict of interest statement
Competing Interests
The remaining authors declare no competing interests.
"V体育官网入口" Figures









VSports最新版本 - References
-
- Stoeckius M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). - PMC (V体育官网入口) - PubMed
-
- Swanson E. et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. Elife 10, (2021). - PMC (V体育官网入口) - PubMed
Methods References
-
- Bujalowski W. & Lohman TM Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry 25, 7799–7802 (1986). - PubMed
-
- Lohman TM & Overman LB Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem 260, 3594–3603 (1985). - V体育官网入口 - PubMed
-
- Reddy MS, Guhan N. & Muniyappa K. Characterization of single-stranded DNA-binding proteins from Mycobacteria. The carboxyl-terminal of domain of SSB is essential for stable association with its cognate RecA protein. J. Biol. Chem 276, 45959–45968 (2001). - PubMed (V体育官网入口)
-
- Parks Benjamin. GreenleafLab/matcha. "V体育2025版" https://github.com/GreenleafLab/matcha.
Publication types
- Actions (V体育官网)
MeSH terms
- Actions (VSports)
"V体育官网入口" Substances
Grants and funding
- U54 HG010426/HG/NHGRI NIH HHS/United States (V体育安卓版)
- R01 HG008140/HG/NHGRI NIH HHS/United States
- U01 DK127419/DK/NIDDK NIH HHS/United States
- U19 AI057266/AI/NIAID NIH HHS/United States
- "V体育平台登录" R01 HG009909/HG/NHGRI NIH HHS/United States
- U01 HG011762/HG/NHGRI NIH HHS/United States (V体育官网)
- U01 MH116529/MH/NIMH NIH HHS/United States
- VSports手机版 - P50 HG007735/HG/NHGRI NIH HHS/United States
- "V体育官网入口" UM1 HG009442/HG/NHGRI NIH HHS/United States
- F32 GM135996/GM/NIGMS NIH HHS/United States (VSports app下载)
- UM1 HG011972/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials (VSports注册入口)

