Deep-joint-learning analysis model of single cell transcriptome and open chromatin accessibility data
- PMID: 33200787
- PMCID: PMC8293818
- DOI: 10.1093/bib/bbaa287 (VSports最新版本)
Deep-joint-learning analysis model of single cell transcriptome and open chromatin accessibility data
Abstract
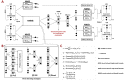
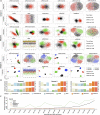
Simultaneous profiling transcriptomic and chromatin accessibility information in the same individual cells offers an unprecedented resolution to understand cell states. However, computationally effective methods for the integration of these inherent sparse and heterogeneous data are lacking. Here, we present a single-cell multimodal variational autoencoder model, which combines three types of joint-learning strategies with a probabilistic Gaussian Mixture Model to learn the joint latent features that accurately represent these multilayer profiles. Studies on both simulated datasets and real datasets demonstrate that it has more preferable capability (i) dissecting cellular heterogeneity in the joint-learning space, (ii) denoising and imputing data and (iii) constructing the association between multilayer omics data, which can be used for understanding transcriptional regulatory mechanisms. VSports手机版.
Keywords: data integration; deep joint-learning model; multimodal variational autoencoder; single-cell multiple omics data. V体育安卓版.
© The Author(s) 2020 V体育ios版. Published by Oxford University Press. .
Figures



References
-
- Mahata B, Zhang XW, Kolodziejczyk AA, et al. . Single-cell RNA sequencing reveals T helper cells synthesizing steroids De Novo to contribute to immune homeostasis. Cell Rep 2014;7:1130–42. - "VSports注册入口" PMC - PubMed
-
- Ziegenhain C, Vieth B, Parekh S, et al. . Comparative analysis of single-cell RNA sequencing methods. Mol Cell 2017;65:631. - PubMed
-
- Kelsey G, Stegle O, Reik W. Single-cell epigenomics: recording the past and predicting the future. Science 2017;358:69–75. - PubMed
V体育官网 - Publication types
MeSH terms (V体育安卓版)
- VSports - Actions
- Actions (V体育2025版)
- Actions (V体育官网)
- "V体育2025版" Actions
- VSports在线直播 - Actions
- Actions (VSports注册入口)
- "V体育2025版" Actions

