VSports手机版 - Frizzled-7 Identifies Platinum-Tolerant Ovarian Cancer Cells Susceptible to Ferroptosis
- PMID: 33172933
- PMCID: PMC7855035
- DOI: 10.1158/0008-5472.CAN-20-1488
Frizzled-7 Identifies Platinum-Tolerant Ovarian Cancer Cells Susceptible to Ferroptosis (V体育官网)
Abstract
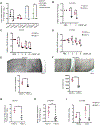
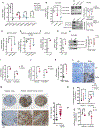
Defining traits of platinum-tolerant cancer cells could expose new treatment vulnerabilities VSports手机版. Here, new markers associated with platinum-tolerant cells and tumors were identified using in vitro and in vivo ovarian cancer models treated repetitively with carboplatin and validated in human specimens. Platinum-tolerant cells and tumors were enriched in ALDH+ cells, formed more spheroids, and expressed increased levels of stemness-related transcription factors compared with parental cells. Additionally, platinum-tolerant cells and tumors exhibited expression of the Wnt receptor Frizzled-7 (FZD7). Knockdown of FZD7 improved sensitivity to platinum, decreased spheroid formation, and delayed tumor initiation. The molecular signature distinguishing FZD7+ from FZD7- cells included epithelial-to-mesenchymal (EMT), stemness, and oxidative phosphorylation-enriched gene sets. Overexpression of FZD7 activated the oncogenic factor Tp63, driving upregulation of glutathione metabolism pathways, including glutathione peroxidase 4 (GPX4), which protected cells from chemotherapy-induced oxidative stress. FZD7+ platinum-tolerant ovarian cancer cells were more sensitive and underwent ferroptosis after treatment with GPX4 inhibitors. FZD7, Tp63, and glutathione metabolism gene sets were strongly correlated in the ovarian cancer Tumor Cancer Genome Atlas (TCGA) database and in residual human ovarian cancer specimens after chemotherapy. These results support the existence of a platinum-tolerant cell population with partial cancer stem cell features, characterized by FZD7 expression and dependent on the FZD7-β-catenin-Tp63-GPX4 pathway for survival. The findings reveal a novel therapeutic vulnerability of platinum-tolerant cancer cells and provide new insight into a potential "persister cancer cell" phenotype. SIGNIFICANCE: Frizzled-7 marks platinum-tolerant cancer cells harboring stemness features and altered glutathione metabolism that depend on GPX4 for survival and are highly susceptible to ferroptosis. .
©2020 American Association for Cancer Research V体育安卓版. .
Conflict of interest statement
Figures





References
-
- Berek JS, Bertelsen K, du Bois A, Brady MF, Carmichael J, Eisenhauer EA, et al. Advanced epithelial ovarian cancer: 1998 consensus statements. Ann Oncol. 1999;10 Suppl 1:87–92. - PubMed
-
- Tummala MK, and McGuire WP. Recurrent ovarian cancer. Clin Adv Hematol Oncol. 2005;3(9):723–36. - PubMed
-
- Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551(7679):247–50. - VSports最新版本 - PMC - PubMed
Publication types
- V体育2025版 - Actions
MeSH terms
- Actions (VSports在线直播)
- Actions (V体育ios版)
- VSports - Actions
- "V体育官网入口" Actions
- "V体育2025版" Actions
- Actions (V体育官网入口)
- "VSports" Actions
- "VSports app下载" Actions
- "V体育平台登录" Actions
- V体育安卓版 - Actions
- Actions (V体育2025版)
- V体育2025版 - Actions
- "V体育2025版" Actions
- Actions (VSports手机版)
- "VSports手机版" Actions
Substances
- "V体育2025版" Actions
- "V体育2025版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

