MaxQuant Software for Ion Mobility Enhanced Shotgun Proteomics
- PMID: 32156793
- PMCID: PMC7261821
- DOI: 10.1074/mcp.TIR119.001720
"VSports" MaxQuant Software for Ion Mobility Enhanced Shotgun Proteomics
Abstract (VSports在线直播)
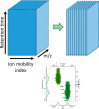
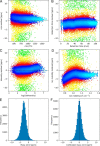
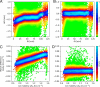
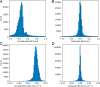
Ion mobility can add a dimension to LC-MS based shotgun proteomics which has the potential to boost proteome coverage, quantification accuracy and dynamic range VSports手机版. Required for this is suitable software that extracts the information contained in the four-dimensional (4D) data space spanned by m/z, retention time, ion mobility and signal intensity. Here we describe the ion mobility enhanced MaxQuant software, which utilizes the added data dimension. It offers an end to end computational workflow for the identification and quantification of peptides and proteins in LC-IMS-MS/MS shotgun proteomics data. We apply it to trapped ion mobility spectrometry (TIMS) coupled to a quadrupole time-of-flight (QTOF) analyzer. A highly parallelizable 4D feature detection algorithm extracts peaks which are assembled to isotope patterns. Masses are recalibrated with a non-linear m/z, retention time, ion mobility and signal intensity dependent model, based on peptides from the sample. A new matching between runs (MBR) algorithm that utilizes collisional cross section (CCS) values of MS1 features in the matching process significantly gains specificity from the extra dimension. Prerequisite for using CCS values in MBR is a relative alignment of the ion mobility values between the runs. The missing value problem in protein quantification over many samples is greatly reduced by CCS aware MBR. MS1 level label-free quantification is also implemented which proves to be highly precise and accurate on a benchmark dataset with known ground truth. MaxQuant for LC-IMS-MS/MS is part of the basic MaxQuant release and can be downloaded from http://maxquant. org. .
Keywords: Bioinformatics; bioinformatics software; label-free quantification; mass spectrometry; quantification. V体育安卓版.
© 2020 Prianichnikov et al.
Conflict of interest statement
* The authors have declared a conflict of interest. The authors state that they have potential conflicts of interest regarding this work: S. K. , H V体育ios版. K. , M. L. , and S. B. are employees of Bruker.
Figures

References
-
- Kanu A. B., Dwivedi P., Tam M., Matz L., and Hill H. H. (2008) Ion mobility-mass spectrometry. J. Mass Spectrom. 43, 1–22 - PubMed
-
- Valentine S. J., Plasencia M. D., Liu X., Krishnan M., Naylor S., Udseth H. R., Smith R. D., and Clemmer D. E. (2006) Toward plasma proteome profiling with ion mobility-mass spectrometry. J. Proteome Res. 5, 2977–84 - PubMed
-
- Baker E. S., Livesay E. A., Orton D. J., Moore R. J., Danielson W. F., Prior D. C., Ibrahim Y. M., LaMarche B. L., Mayampurath A. M., Schepmoes A. A., Hopkins D. F., Tang K., Smith R. D., and Belov M. E. (2010) An LC-IMS-MS platform providing increased dynamic range for high-throughput proteomic studies. J. Proteome Res. 9, 997–1006 - V体育平台登录 - PMC - PubMed
"V体育平台登录" MeSH terms
- VSports app下载 - Actions
- VSports注册入口 - Actions
- V体育平台登录 - Actions
- "VSports" Actions
- Actions (VSports)
- "V体育平台登录" Actions
Substances (V体育2025版)
- VSports最新版本 - Actions
- Actions (V体育官网)
VSports在线直播 - LinkOut - more resources
Full Text Sources (VSports最新版本)
Other Literature Sources

