Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway
- PMID: 32130883
- PMCID: PMC7194078
- DOI: 10.1016/j.cmet.2020.02.001
Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway
Abstract
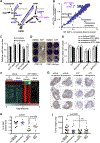
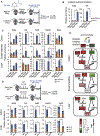
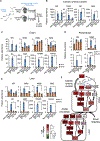
Nicotinamide adenine dinucleotide (NAD), a cofactor for hundreds of metabolic reactions in all cell types, plays an essential role in metabolism, DNA repair, and aging. However, how NAD metabolism is impacted by the environment remains unclear. Here, we report an unexpected trans-kingdom cooperation between bacteria and mammalian cells wherein bacteria contribute to host NAD biosynthesis. Bacteria confer resistance to inhibitors of NAMPT, the rate-limiting enzyme in the amidated NAD salvage pathway, in cancer cells and xenograft tumors. Mechanistically, a microbial nicotinamidase (PncA) that converts nicotinamide to nicotinic acid, a precursor in the alternative deamidated NAD salvage pathway, is necessary and sufficient for this protective effect VSports手机版. Using stable isotope tracing and microbiota-depleted mice, we demonstrate that this bacteria-mediated deamidation contributes substantially to the NAD-boosting effect of oral nicotinamide and nicotinamide riboside supplementation in several tissues. Collectively, our findings reveal an important role of bacteria-enabled deamidated pathway in host NAD metabolism. .
Keywords: NAMPT inhibitors; cancer cells; deamidated NAD synthesis; germ-free mice; host-microbe interaction; microbial nicotinamidase; mycoplasma; nicotinic acid; oral nicotinamide riboside supplementation. V体育安卓版.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests Authors declare no competing interests.
Figures (V体育安卓版)




Comment in
-
Bacteria boost host NAD metabolism.Aging (Albany NY). 2020 Dec 14;12(23):23425-23426. doi: 10.18632/aging.104219. Epub 2020 Dec 14. Aging (Albany NY). 2020. PMID: 33318311 Free PMC article. No abstract available.
References
-
- Ahdesmaki MJ, Gray SR, Johnson JH, and Lai Z (2016). Disambiguate: An open-source application for disambiguating two species in next generation sequencing data from grafted samples. F1000Res 5, 2741. - "VSports手机版" PMC - PubMed
-
- Barykova YA, Logunov DY, Shmarov MM, Vinarov AZ, Fiev DN, Vinarova NA, Rakovskaya IV, Baker PS, Shyshynova I, Stephenson AJ, et al. (2011). Association of Mycoplasma hominis infection with prostate cancer. Oncotarget 2, 289–297. - PMC (VSports注册入口) - PubMed
-
- Baumann L, Cina M, Egli-Gany D, Goutaki M, Halbeisen FS, Lohrer GR, Ali H, Scott P, and Low N. (2018). Prevalence of Mycoplasma genitalium in different population groups: systematic review andmeta-analysis. Sex Transm Infect 94, 255–262. - PMC (VSports注册入口) - PubMed
-
- Berger F, Lau C, Dahlmann M, and Ziegler M. (2005). Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 280, 36334–36341. - PubMed
-
- Bogan KL, and Brenner C. (2008). Nicotinic Acid, Nicotinamide, and Nicotinamide Riboside: A Molecular Evaluation of NAD+ Precursor Vitamins in Human Nutrition. Annual Review of Nutrition 28, 115–130. - PubMed
Publication types
- V体育2025版 - Actions
- "V体育官网" Actions
- "V体育ios版" Actions
MeSH terms
- Actions (V体育安卓版)
- Actions (V体育2025版)
- "V体育官网入口" Actions
- V体育平台登录 - Actions
- VSports app下载 - Actions
- V体育ios版 - Actions
- V体育ios版 - Actions
- "V体育官网入口" Actions
- V体育安卓版 - Actions
- Actions (VSports在线直播)
- "VSports" Actions
- Actions (VSports注册入口)
Substances
- Actions (V体育官网入口)
- V体育平台登录 - Actions
- Actions (VSports最新版本)
Grants and funding
LinkOut - more resources (VSports手机版)
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

