A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1
- PMID: 31470874
- PMCID: PMC6716823
- DOI: 10.1186/s12943-019-1056-5
V体育2025版 - A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1
Erratum in (VSports在线直播)
-
Correction to: A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1.Mol Cancer. 2019 Oct 29;18(1):149. doi: 10.1186/s12943-019-1083-2. Mol Cancer. 2019. PMID: 31660951 Free PMC article.
-
Correction: A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1.Mol Cancer. 2022 Jun 7;21(1):124. doi: 10.1186/s12943-022-01584-y. Mol Cancer. 2022. PMID: 35672742 Free PMC article. No abstract available.
Abstract
Background: The RTK/PI3K/AKT pathway plays key roles in the development and progression of many cancers, including GBM. As a regulatory molecule and a potential drug target, the oncogenic role of AKT has been substantially studied. Three isoforms of AKT have been identified, including AKT1, AKT2 and AKT3, but their individual functions in GBM remain controversial VSports手机版. Moreover, it is not known if there are more AKT alternative splicing variants. .
Methods: High-throughput RNA sequencing and quantitative reverse transcription-PCR were used to identify the differentially expressed circRNAs in GBM samples and in paired normal tissues. High throughput RNA sequencing was used to identify circ-AKT3 regulated signaling pathways. Mass spectrometry, western blotting and immunofluorescence staining analyses were used to validate AKT3-174aa expression. The tumor suppressive role of AKT3-174aa was validated in vitro and in vivo. The competing interaction between AKT3-174aa and p-PDK1 was investigated by mass spectrometry and immunoprecipitation analyses V体育安卓版. .
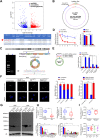
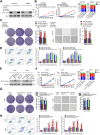
Results: Circ-AKT3 is a previously uncharacterized AKT transcript variant. Circ-AKT3 is expressed at low levels in GBM tissues compared with the expression in paired adjacent normal brain tissues. Circ-AKT3 encodes a 174 amino acid (aa) novel protein, which we named AKT3-174aa, by utilizing overlapping start-stop codons V体育ios版. AKT3-174aa overexpression decreased the cell proliferation, radiation resistance and in vivo tumorigenicity of GBM cells, while the knockdown of circ-AKT3 enhanced the malignant phenotypes of astrocytoma cells. AKT3-174aa competitively interacts with phosphorylated PDK1, reduces AKT-thr308 phosphorylation, and plays a negative regulatory role in modulating the PI3K/AKT signal intensity. .
Conclusions: Our data indicate that the impaired circRNA expression of the AKT3 gene contributes to GBM tumorigenesis, and our data corroborate the hypothesis that restoring AKT3-174aa while inhibiting activated AKT may provide more benefits for certain GBM patients. VSports最新版本.
Keywords: AKT3; Glioblastoma; PDK1; circRNA. V体育平台登录.
Conflict of interest statement
The authors declare that they have no competing interests.
"VSports注册入口" Figures





References
-
- Chin YR, Yuan X, Balk SP, Toker A. PTEN-deficient tumors depend on AKT2 for maintenance and survival. Cancer Discov. 2014;4:942–955. doi: 10.1158/2159-8290.CD-13-0873. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

