Resistance to paclitaxel is associated with a variant of the gene BCL2 in multiple tumor types (VSports)
- PMID: 31044156
- PMCID: "V体育安卓版" PMC6478919
- DOI: 10.1038/s41698-019-0084-3
Resistance to paclitaxel is associated with a variant of the gene BCL2 in multiple tumor types
Abstract (V体育2025版)
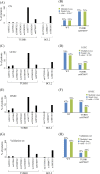
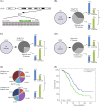
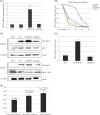
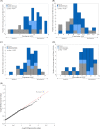
Paclitaxel, the most commonly used form of chemotherapy, is utilized in curative protocols in different types of cancer. The response to treatment differs among patients. Biological interpretation of a mechanism to explain this personalized response is still unavailable. Since paclitaxel is known to target BCL2 and TUBB1, we used pan-cancer genomic data from hundreds of patients to show that a single-nucleotide variant in the BCL2 sequence can predict a patient's response to paclitaxel. Here, we show a connection between this BCL2 genomic variant, its transcript structure, and protein abundance. We demonstrate these findings in silico, in vitro, in formalin-fixed paraffin-embedded (FFPE) tissue, and in patient lymphocytes. We show that tumors with the specific variant are more resistant to paclitaxel. We also show that tumor and normal cells with the variant express higher levels of BCL2 protein, a phenomenon that we validated in an independent cohort of patients. Our results indicate BCL2 sequence variations as determinants of chemotherapy resistance. The knowledge of individual BCL2 genomic sequences prior to the choice of chemotherapy may improve patient survival VSports手机版. The current work also demonstrates the benefit of community-wide, integrative omics data sources combined with in-lab experimentation and validation sets. .
Keywords: Cancer genomics; High-throughput screening; Molecular biology V体育安卓版. .
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Links M, Brown R. Clinical relevance of the molecular mechanisms of resistance to anti-cancer drugs. Expert Rev. Mol. Med. 1999;1999:1–21. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

