RPA1 binding to NRF2 switches ARE-dependent transcriptional activation to ARE-NRE-dependent repression
- PMID: 30309964
- PMCID: PMC6217430
- DOI: 10.1073/pnas.1812125115
RPA1 binding to NRF2 switches ARE-dependent transcriptional activation to ARE-NRE-dependent repression
"VSports在线直播" Abstract
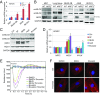
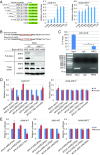
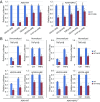
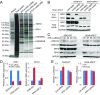
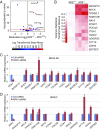
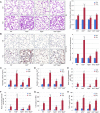
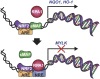
NRF2 regulates cellular redox homeostasis, metabolic balance, and proteostasis by forming a dimer with small musculoaponeurotic fibrosarcoma proteins (sMAFs) and binding to antioxidant response elements (AREs) to activate target gene transcription. In contrast, NRF2-ARE-dependent transcriptional repression is unreported. Here, we describe NRF2-mediated gene repression via a specific seven-nucleotide sequence flanking the ARE, which we term the NRF2-replication protein A1 (RPA1) element (NRE). Mechanistically, RPA1 competes with sMAF for NRF2 binding, followed by interaction of NRF2-RPA1 with the ARE-NRE and eduction of promoter activity. Genome-wide in silico and RNA-seq analyses revealed this NRF2-RPA1-ARE-NRE complex mediates negative regulation of many genes with diverse functions, indicating that this mechanism is a fundamental cellular process VSports手机版. Notably, repression of MYLK, which encodes the nonmuscle myosin light chain kinase, by the NRF2-RPA1-ARE-NRE complex disrupts vascular integrity in preclinical inflammatory lung injury models, illustrating the translational significance of NRF2-mediated transcriptional repression. Our findings reveal a gene-suppressive function of NRF2 and a subset of negatively regulated NRF2 target genes, underscoring the broad impact of NRF2 in physiological and pathological settings. .
Keywords: MYLK/MLCK; NRF2; RPA1; acute lung injury; transcriptional regulation V体育安卓版. .
Conflict of interest statement
The authors declare no conflict of interest.
VSports在线直播 - Figures

"VSports手机版" References
-
- Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. - PMC (V体育安卓版) - PubMed
-
- Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. - V体育平台登录 - PubMed
-
- Mitsuishi Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. - PubMed
Publication types
MeSH terms
- Actions (V体育官网入口)
- "V体育2025版" Actions
- "VSports在线直播" Actions
- VSports手机版 - Actions
- Actions (VSports app下载)
- Actions (VSports手机版)
V体育2025版 - Substances
- "V体育平台登录" Actions
Grants and funding
LinkOut - more resources (VSports最新版本)
Full Text Sources
Molecular Biology Databases

