Refined RIP-seq protocol for epitranscriptome analysis with low input materials
- PMID: 30212448
- PMCID: "V体育2025版" PMC6136692
- DOI: 10.1371/journal.pbio.2006092
V体育2025版 - Refined RIP-seq protocol for epitranscriptome analysis with low input materials
Abstract
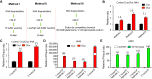
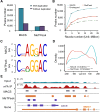
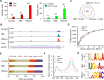
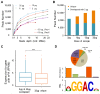
N6-Methyladenosine (m6A) accounts for approximately 0. 2% to 0. 6% of all adenosine in mammalian mRNA, representing the most abundant internal mRNA modifications. m6A RNA immunoprecipitation followed by high-throughput sequencing (MeRIP-seq) is a powerful technique to map the m6A location transcriptome-wide. However, this method typically requires 300 μg of total RNA, which limits its application to patient tumors. In this study, we present a refined m6A MeRIP-seq protocol and analysis pipeline that can be applied to profile low-input RNA samples from patient tumors. We optimized the key parameters of m6A MeRIP-seq, including the starting amount of RNA, RNA fragmentation, antibody selection, MeRIP washing/elution conditions, methods for RNA library construction, and the bioinformatics analysis pipeline. With the optimized immunoprecipitation (IP) conditions and a postamplification rRNA depletion strategy, we were able to profile the m6A epitranscriptome using 500 ng of total RNA. We identified approximately 12,000 m6A peaks with a high signal-to-noise (S/N) ratio from 2 lung adenocarcinoma (ADC) patient tumors. Through integrative analysis of the transcriptome, m6A epitranscriptome, and proteome data in the same patient tumors, we identified dynamics at the m6A level that account for the discordance between mRNA and protein levels in these tumors. The refined m6A MeRIP-seq method is suitable for m6A epitranscriptome profiling in a limited amount of patient tumors, setting the ground for unraveling the dynamics of the m6A epitranscriptome and the underlying mechanisms in clinical settings. VSports手机版.
Conflict of interest statement
The method is subjected to a University Health Network patent application with Shiyan Wang, Yong Zeng, and Housheng He as inventors.
Figures

References
-
- Roignant J-Y, Soller M. m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017;33: 380–390. 10.1016/j.tig.2017.04.003 - DOI (VSports最新版本) - PubMed
-
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. National Academy of Sciences; 1974;71: 3971–3975. - V体育2025版 - PMC - PubMed
Publication types
MeSH terms
- V体育官网入口 - Actions
- "VSports最新版本" Actions
- Actions (V体育平台登录)
- "V体育官网入口" Actions
- V体育安卓版 - Actions
- "VSports在线直播" Actions
Substances
- "V体育ios版" Actions
- Actions (VSports app下载)
- "VSports" Actions
V体育ios版 - LinkOut - more resources
Full Text Sources
Other Literature Sources
"VSports注册入口" Molecular Biology Databases
Miscellaneous

