m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer (VSports注册入口)
- PMID: 30154548
- PMCID: PMC6245953
- DOI: 10.1038/s41556-018-0174-4
m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
Abstract (VSports注册入口)
N6-methyladenosine (m6A) messenger RNA methylation is a gene regulatory mechanism affecting cell differentiation and proliferation in development and cancer. To study the roles of m6A mRNA methylation in cell proliferation and tumorigenicity, we investigated human endometrial cancer in which a hotspot R298P mutation is present in a key component of the methyltransferase complex (METTL14). We found that about 70% of endometrial tumours exhibit reductions in m6A methylation that are probably due to either this METTL14 mutation or reduced expression of METTL3, another component of the methyltransferase complex. These changes lead to increased proliferation and tumorigenicity of endometrial cancer cells, likely through activation of the AKT pathway. Reductions in m6A methylation lead to decreased expression of the negative AKT regulator PHLPP2 and increased expression of the positive AKT regulator mTORC2. Together, these results reveal reduced m6A mRNA methylation as an oncogenic mechanism in endometrial cancer and identify m6A methylation as a regulator of AKT signalling. VSports手机版.
"V体育平台登录" Conflict of interest statement
COMPETING FINANCIAL INTERESTS
C. H. is a scientific founder of Accent Therapeutics and a member of its scientific advisory board V体育安卓版. All other authors declare no competing financial interests.
Figures (V体育ios版)

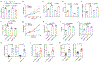


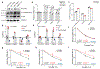
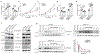

References (V体育官网)
-
- Liu N & Pan T N6-methyladenosine–encoded epitranscriptomics. Nat Struct Mol Biol 23, 98–102 (2016). - PubMed
-
- Zhao BS, Roundtree IA & He C Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18, 31–42 (2017). - PMC (V体育ios版) - PubMed
-
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P & Rottman F Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem 269, 17697–17704 (1994). - PubMed
Publication types
- Actions (V体育官网)
MeSH terms
- Actions (VSports最新版本)
- Actions (V体育ios版)
- "VSports注册入口" Actions
- "V体育2025版" Actions
- V体育官网入口 - Actions
- "VSports" Actions
- V体育官网 - Actions
- Actions (V体育平台登录)
- V体育2025版 - Actions
- "VSports app下载" Actions
- Actions (VSports最新版本)
- Actions (V体育ios版)
Substances
- Actions (V体育安卓版)
- Actions (V体育ios版)
- "VSports在线直播" Actions
- "V体育官网入口" Actions
- "VSports app下载" Actions
- Actions (V体育平台登录)
V体育安卓版 - Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

