"V体育安卓版" The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5'-3' exoribonuclease XRN1
- PMID: 29970596
- PMCID: V体育官网 - PMC6140455
- DOI: 10.1261/rna.064238.117
The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5'-3' exoribonuclease XRN1
Abstract (V体育平台登录)
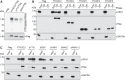
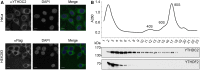
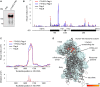
N6-methyladenosine (m6A) modifications in RNAs play important roles in regulating many different aspects of gene expression. While m6As can have direct effects on the structure, maturation, or translation of mRNAs, such modifications can also influence the fate of RNAs via proteins termed "readers" that specifically recognize and bind modified nucleotides. Several YTH domain-containing proteins have been identified as m6A readers that regulate the splicing, translation, or stability of specific mRNAs. In contrast to the other YTH domain-containing proteins, YTHDC2 has several defined domains and here, we have analyzed the contribution of these domains to the RNA and protein interactions of YTHDC2. The YTH domain of YTHDC2 preferentially binds m6A-containing RNAs via a conserved hydrophobic pocket, whereas the ankyrin repeats mediate an RNA-independent interaction with the 5'-3' exoribonuclease XRN1. We show that the YTH and R3H domains contribute to the binding of YTHDC2 to cellular RNAs, and using crosslinking and analysis of cDNA (CRAC), we reveal that YTHDC2 interacts with the small ribosomal subunit in close proximity to the mRNA entry/exit sites. YTHDC2 was recently found to promote a "fast-track" expression program for specific mRNAs, and our data suggest that YTHDC2 accomplishes this by recruitment of the RNA degradation machinery to regulate the stability of m6A-containing mRNAs and by utilizing its distinct RNA-binding domains to bridge interactions between m6A-containing mRNAs and the ribosomes to facilitate their efficient translation. VSports手机版.
Keywords: N6-methyladenosine (m6A); RNA modification; YTH domain; exoribonuclease; ribosome; translation V体育安卓版. .
© 2018 Kretschmer et al. ; Published by Cold Spring Harbor Laboratory Press for the RNA Society V体育ios版. .
Figures

VSports app下载 - References
-
- Adams JM, Cory S. 1975. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 255: 28–33. - "V体育2025版" PubMed
-
- Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. 2015. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162: 1299–1308. - V体育平台登录 - PMC - PubMed
-
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85. - PubMed
Publication types
- V体育官网入口 - Actions
MeSH terms
- "VSports最新版本" Actions
- "V体育官网" Actions
- V体育官网入口 - Actions
- VSports最新版本 - Actions
- VSports在线直播 - Actions
- V体育官网入口 - Actions
- "VSports最新版本" Actions
- Actions (V体育2025版)
- V体育安卓版 - Actions
- "V体育官网" Actions
- "VSports app下载" Actions
VSports在线直播 - Substances
- V体育安卓版 - Actions
- "V体育平台登录" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
