Kaposi's Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N6-Adenosine Methylation To Promote Lytic Replication
- PMID: 28592530
- PMCID: "VSports" PMC5533915
- DOI: 10.1128/JVI.00466-17
Kaposi's Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N6-Adenosine Methylation To Promote Lytic Replication
Abstract
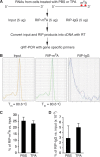
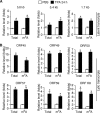
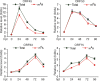
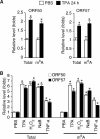
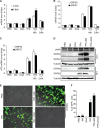
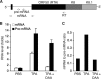
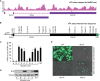
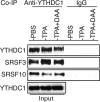
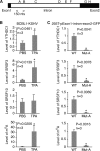
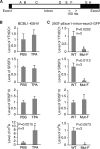
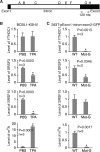
N6-adenosine methylation (m6A) is the most common posttranscriptional RNA modification in mammalian cells. We found that most transcripts encoded by the Kaposi's sarcoma-associated herpesvirus (KSHV) genome undergo m6A modification. The levels of m6A-modified mRNAs increased substantially upon stimulation for lytic replication. The blockage of m6A inhibited splicing of the pre-mRNA encoding the replication transcription activator (RTA), a key KSHV lytic switch protein, and halted viral lytic replication. We identified several m6A sites in RTA pre-mRNA crucial for splicing through interactions with YTH domain containing 1 (YTHDC1), an m6A nuclear reader protein, in conjunction with serine/arginine-rich splicing factor 3 (SRSF3) and SRSF10 VSports手机版. Interestingly, RTA induced m6A and enhanced its own pre-mRNA splicing. Our results not only demonstrate an essential role of m6A in regulating RTA pre-mRNA splicing but also suggest that KSHV has evolved a mechanism to manipulate the host m6A machinery to its advantage in promoting lytic replication. IMPORTANCE KSHV productive lytic replication plays a pivotal role in the initiation and progression of Kaposi's sarcoma tumors. Previous studies suggested that the KSHV switch from latency to lytic replication is primarily controlled at the chromatin level through histone and DNA modifications. The present work reports for the first time that KSHV genome-encoded mRNAs undergo m6A modification, which represents a new mechanism at the posttranscriptional level in the control of viral replication. .
Keywords: KSHV; N6-adenosine methylation; RNA splicing; lytic replication V体育安卓版. .
Copyright © 2017 American Society for Microbiology. V体育ios版.
Figures




"V体育ios版" References
-
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149:1635–1646. doi:10.1016/j.cell.2012.05.003. - VSports app下载 - DOI - PMC - PubMed
-
- Adams JM, Cory S. 1975. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 255:28–33. doi:10.1038/255028a0. - "V体育官网入口" DOI - PubMed
-
- Desrosiers R, Friderici K, Rottman F. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 71:3971–3975. doi:10.1073/pnas.71.10.3971. - DOI (V体育官网入口) - PMC - PubMed
Publication types
MeSH terms
- Actions (VSports最新版本)
- Actions (VSports手机版)
- Actions (V体育平台登录)
- Actions (VSports在线直播)
Substances
- "V体育ios版" Actions
- "VSports" Actions
- VSports注册入口 - Actions
"V体育2025版" Grants and funding
LinkOut - more resources
Full Text Sources
"V体育平台登录" Other Literature Sources
Miscellaneous

