CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome
- PMID: 28369033
- PMCID: PMC5462860
- DOI: 10.1038/nbt.3853
CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome
Abstract
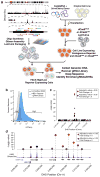
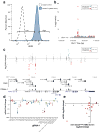
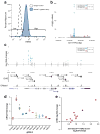
Large genome-mapping consortia and thousands of genome-wide association studies have identified non-protein-coding elements in the genome as having a central role in various biological processes. However, decoding the functions of the millions of putative regulatory elements discovered in these studies remains challenging. CRISPR-Cas9-based epigenome editing technologies have enabled precise perturbation of the activity of specific regulatory elements. Here we describe CRISPR-Cas9-based epigenomic regulatory element screening (CERES) for improved high-throughput screening of regulatory element activity in the native genomic context. Using dCas9KRAB repressor and dCas9p300 activator constructs and lentiviral single guide RNA libraries to target DNase I hypersensitive sites surrounding a gene of interest, we carried out both loss- and gain-of-function screens to identify regulatory elements for the β-globin and HER2 loci in human cells VSports手机版. CERES readily identified known and previously unidentified regulatory elements, some of which were dependent on cell type or direction of perturbation. This technology allows the high-throughput functional annotation of putative regulatory elements in their native chromosomal context. .
Conflict of interest statement
TSK, JBB, IBH, GEC, TER, and CAG are named inventors on patent applications related to genome engineering. TSK, GEC, TER, and CAG are founders of Element Genomics V体育安卓版.
Figures

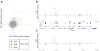
"V体育2025版" References
-
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. - PMC - PubMed
-
- Hindorff LA, Junkins HA, Mehta JP, Manolio TA. OPG: Catalog Published Genome-Wide Assoc Studies. 2009.
-
- Arnold CD, Gerlach D, Stelzer C, Boryń ŁM, Rath M, Stark A. Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq. Science. 2013;339:1074–1077. - PubMed
Publication types (VSports)
- "VSports" Actions
- V体育平台登录 - Actions
MeSH terms
- VSports注册入口 - Actions
- "VSports" Actions
- VSports手机版 - Actions
- Actions (VSports在线直播)
- "VSports" Actions
Substances
- "V体育安卓版" Actions
Grants and funding
V体育2025版 - LinkOut - more resources
Full Text Sources
Other Literature Sources
V体育平台登录 - Molecular Biology Databases
Research Materials
Miscellaneous

