MicroRNA-1908-5p contributes to the oncogenic function of the splicing factor SRSF3
- PMID: 28039456
- PMCID: PMC5352405
- DOI: 10.18632/oncotarget.14184
MicroRNA-1908-5p contributes to the oncogenic function of the splicing factor SRSF3
Abstract
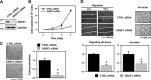
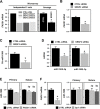
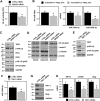
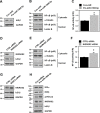
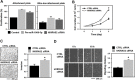
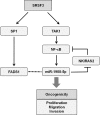
Serine/arginine (SR)-rich proteins that contain RS domains and SR repeats have diverse cellular functions including transcription, polyadenylation, translation, and RNA export. The splicing factor SRSF3, also termed SRp20, is the smallest member of the SR protein family and is a known proto-oncogene. Although it is implicated in the malignant phenotypes of various cancer cells, the molecular mechanism underlying SRSF3-mediated cancer progression is still obscure VSports手机版. We investigated here the oncogenic functions of SRSF3 in osteosarcoma U2OS cells. Knockdown of SRSF3 inhibited proliferation, clonogenicity, and metastatic potential including migration and invasion. It also decreased the level of miR-1908 independent of its host gene FADS1. Although FADS1 was not associated with SRSF3-mediated malignant properties, overexpression of miR-1908-5p increased cell proliferation, migration, and invasion, suggesting that miR-1908-5p is responsible for the oncogenic functions of SRSF3. Knockdown of SRSF3 decreased the expression of miR-1908-5p by inhibiting transactivation of NF-κB. We observed that miR-1908-5p downregulated NF-κB inhibitor interacting Ras-like 2 (NKIRAS2), a negative regulator of the NF-κB pathway by directly binding to the 3'UTR of NKIRAS2 mRNA. Consistent with overexpression of miR-1908-5p, knockdown of NKIRAS2 diminished the expression level of IκB-β and provoked translocation of NF-κB into the nucleus where it transcriptionally activates its target genes including miR-1908-5p expression, thus elevating the proliferation and metastatic potential. Taken together, our results demonstrate that SRSF3 confers the malignant characteristics on cancer cells via the SRSF3/miR-1908-5p/NKIRAS2 axis. .
Keywords: FADS1; NF-κB; NKIRAS2; SRSF3; miR-1908-5p V体育安卓版. .
Conflict of interest statement
The authors declare no competing financial interests.
"V体育官网入口" Figures


References
-
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–47. - PubMed
MeSH terms
- "V体育ios版" Actions
- "V体育ios版" Actions
- Actions (VSports)
- "V体育官网入口" Actions
- VSports注册入口 - Actions
- "VSports app下载" Actions
- V体育安卓版 - Actions
- "V体育ios版" Actions
- "VSports最新版本" Actions
- V体育ios版 - Actions
- Actions (V体育安卓版)
- "V体育平台登录" Actions
- "V体育官网入口" Actions
- Actions (VSports在线直播)
Substances
- VSports手机版 - Actions
- "V体育官网" Actions
- V体育安卓版 - Actions
- Actions (VSports)
- "VSports最新版本" Actions
- Actions (V体育平台登录)
LinkOut - more resources
Full Text Sources
Other Literature Sources
VSports - Medical
V体育2025版 - Research Materials
Miscellaneous

