VSports最新版本 - The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9
- PMID: 27325748
- PMCID: PMC5062969
- DOI: 10.1093/nar/gkw564
The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9
Abstract
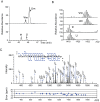
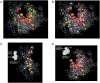
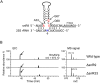
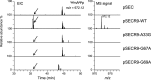
We present the complete chemical structures of the rRNAs from the eukaryotic model organism, Saccharomyces cerevisiae The final structures, as determined with mass spectrometry-based methodology that includes a stable isotope-labelled, non-modified reference RNA, contain 112 sites with 12 different post-transcriptional modifications, including a previously unidentified pseudouridine at position 2345 in 25S rRNA. Quantitative mass spectrometry-based stoichiometric analysis of the different modifications at each site indicated that 94 sites were almost fully modified, whereas the remaining 18 sites were modified to a lesser extent. Superimposed three-dimensional modification maps for S. cerevisiae and Schizosaccharomyces pombe rRNAs confirmed that most of the modified nucleotides are located in functionally important interior regions of the ribosomes. We identified snR9 as the snoRNA responsible for pseudouridylation of U2345 and showed that this pseudouridylation occurs co-transcriptionally and competitively with 2'-O-methylation of U2345 VSports手机版. This study ends the uncertainty concerning whether all modified nucleotides in S. cerevisiae rRNAs have been identified and provides a resource for future structural, functional and biogenesis studies of the eukaryotic ribosome. .
© The Author(s) 2016 V体育安卓版. Published by Oxford University Press on behalf of Nucleic Acids Research. .
Figures (V体育安卓版)
References
-
- Yusupova G., Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014;83:467–486. - "VSports手机版" PubMed
-
- Voigts-Hoffmann F., Klinge S., Ban N. Structural insights into eukaryotic ribosomes and the initiation of translation. Curr. Opin. Struct. Biol. 2012;22:768–777. - PubMed
-
- Jenner L., Melnikov S., Garreau de Loubresse N., Ben-Shem A., Iskakova M., Urzhumtsev A., Meskauskas A., Dinman J., Yusupova G., Yusupov M. Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 2012;22:759–767. - PubMed (V体育安卓版)
-
- Klinge S., Voigts-Hoffmann F., Leibundgut M., Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci. 2012;37:189–198. - PubMed
MeSH terms
- "V体育平台登录" Actions
- Actions (V体育安卓版)
- "VSports手机版" Actions
- "VSports手机版" Actions
- Actions (VSports手机版)
Substances
- V体育2025版 - Actions
- Actions (V体育平台登录)
- "V体育ios版" Actions
- Actions (V体育ios版)
LinkOut - more resources
V体育ios版 - Full Text Sources
Other Literature Sources (V体育平台登录)
Molecular Biology Databases

