Programmable RNA Tracking in Live Cells with CRISPR/Cas9
- PMID: 26997482
- PMCID: PMC4826288 (V体育2025版)
- DOI: 10.1016/j.cell.2016.02.054 (VSports)
"V体育官网" Programmable RNA Tracking in Live Cells with CRISPR/Cas9
Abstract
RNA-programmed genome editing using CRISPR/Cas9 from Streptococcus pyogenes has enabled rapid and accessible alteration of specific genomic loci in many organisms. A flexible means to target RNA would allow alteration and imaging of endogenous RNA transcripts analogous to CRISPR/Cas-based genomic tools, but most RNA targeting methods rely on incorporation of exogenous tags VSports手机版. Here, we demonstrate that nuclease-inactive S. pyogenes CRISPR/Cas9 can bind RNA in a nucleic-acid-programmed manner and allow endogenous RNA tracking in living cells. We show that nuclear-localized RNA-targeting Cas9 (RCas9) is exported to the cytoplasm only in the presence of sgRNAs targeting mRNA and observe accumulation of ACTB, CCNA2, and TFRC mRNAs in RNA granules that correlate with fluorescence in situ hybridization. We also demonstrate time-resolved measurements of ACTB mRNA trafficking to stress granules. Our results establish RCas9 as a means to track RNA in living cells in a programmable manner without genetically encoded tags. .
Copyright © 2016 Elsevier Inc V体育安卓版. All rights reserved. .
Figures

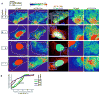
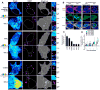
Comment in
-
Technique: Transcript tracking by CRISPR.Nat Rev Genet. 2016 May;17(5):254-5. doi: 10.1038/nrg.2016.41. Epub 2016 Mar 30. Nat Rev Genet. 2016. PMID: 27040488 No abstract available.
References
-
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. - PubMed
-
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. - PubMed
-
- Chou HH, Hsia AP, Mooney DL, Schnable PS. Picky: oligo microarray design for large genomes. Bioinformatics. 2004;20:2893–2902. - "V体育2025版" PubMed
Publication types
- V体育官网入口 - Actions
- V体育官网入口 - Actions
"V体育平台登录" MeSH terms
- V体育ios版 - Actions
- VSports注册入口 - Actions
- Actions (VSports在线直播)
- VSports app下载 - Actions
- "V体育官网" Actions
Substances
Grants and funding
LinkOut - more resources
VSports手机版 - Full Text Sources
V体育安卓版 - Other Literature Sources
Research Materials
Miscellaneous

