Structures of the CDK12/CycK complex with AMP-PNP reveal a flexible C-terminal kinase extension important for ATP binding (VSports)
- PMID: 26597175
- PMCID: PMC4656997
- DOI: 10.1038/srep17122
Structures of the CDK12/CycK complex with AMP-PNP reveal a flexible C-terminal kinase extension important for ATP binding
Abstract
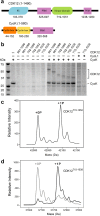
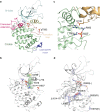
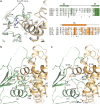
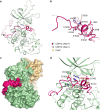
Cyclin-dependent kinase 12 (CDK12) promotes transcriptional elongation by phosphorylation of the RNA polymerase II C-terminal domain (CTD). Structure-function studies show that this activity is dependent on a C-terminal kinase extension, as well as the binding of cyclin K (CycK). To better define these interactions we determined the crystal structure of the human CDK12/CycK complex with and without the kinase extension in the presence of AMP-PNP. The structures revealed novel features for a CDK, including a large β4-β5 loop insertion that contributes to the N-lobe interaction with the cyclin VSports手机版. We also observed two different conformations of the C-terminal kinase extension that effectively open and close the ATP pocket. Most notably, bound AMP-PNP was only observed when trapped in the closed state. Truncation of this C-terminal structure also diminished AMP-PNP binding, as well as the catalytic activity of the CDK12/CycK complex. Further kinetic measurements showed that the full length CDK12/CycK complex was significantly more active than the two crystallised constructs suggesting a critical role for additional domains. Overall, these results demonstrate the intrinsic flexibility of the C-terminal extension in CDK12 and highlight its importance for both ATP binding and kinase activity. .
Figures
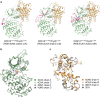


References
-
- Lim S. & Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 140, 3079–93 (2013). - PubMed
-
- Mikolcevic P. et al. Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Mol Cell Biol 32, 868–79 (2012). - VSports app下载 - PMC - PubMed
-
- Davidson G. et al. Cell cycle control of wnt receptor activation. Dev Cell 17, 788–99 (2009). - PubMed
-
- Mikolcevic P., Rainer J. & Geley S. Orphan kinases turn eccentric: a new class of cyclin Y-activated, membrane-targeted CDKs. Cell Cycle 11, 3758–68 (2012). - V体育官网 - PMC - PubMed
Publication types
- "VSports最新版本" Actions
MeSH terms
- VSports在线直播 - Actions
- VSports在线直播 - Actions
- V体育安卓版 - Actions
- VSports手机版 - Actions
- VSports最新版本 - Actions
Substances (V体育官网入口)
- "V体育安卓版" Actions
- "V体育官网" Actions
- Actions (V体育2025版)
Associated data
- Actions
- VSports - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

