Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta
- PMID: 25866491
- PMCID: V体育平台登录 - PMC4390092
- DOI: 10.1021/ct500864r
Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta
Abstract
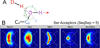
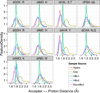
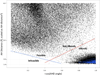
Interactions between polar atoms are challenging to model because at very short ranges they form hydrogen bonds (H-bonds) that are partially covalent in character and exhibit strong orientation preferences; at longer ranges the orientation preferences are lost, but significant electrostatic interactions between charged and partially charged atoms remain VSports手机版. To simultaneously model these two types of behavior, we refined an orientation dependent model of hydrogen bonds [Kortemme et al. J. Mol. Biol. 2003, 326, 1239] used by the molecular modeling program Rosetta and then combined it with a distance-dependent Coulomb model of electrostatics. The functional form of the H-bond potential is physically motivated and parameters are fit so that H-bond geometries that Rosetta generates closely resemble H-bond geometries in high-resolution crystal structures. The combined potentials improve performance in a variety of scientific benchmarks including decoy discrimination, side chain prediction, and native sequence recovery in protein design simulations and establishes a new standard energy function for Rosetta. .
Figures (V体育平台登录)

References
-
- Baker EN, Hubbard RE. Prog. Biophys. molec. Biol. 1984;44:97. - PubMed
-
- Fersht AR, Shi JP, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. Nature. 1985;314:235. - PubMed
-
- Müller-Dethlefs K, Hobza P. Chem. Rev. 2000;100:143. - PubMed
-
- Morozov AV, Kortemme T. Adv. Protein Chem. 2005;72:1. - PubMed
-
- Forrest R, Honig B. Proteins. 2005;61:296. - "VSports最新版本" PubMed
Publication types
MeSH terms
- "VSports在线直播" Actions
- "V体育安卓版" Actions
- "VSports最新版本" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources (VSports app下载)
