The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo
- PMID: 25843629
- PMCID: PMC6922011
- DOI: 10.1016/j.cell.2015.02.043
The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo
Abstract
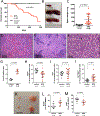
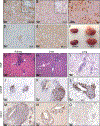
Research over the past decade has suggested important roles for pseudogenes in physiology and disease. In vitro experiments demonstrated that pseudogenes contribute to cell transformation through several mechanisms. However, in vivo evidence for a causal role of pseudogenes in cancer development is lacking VSports手机版. Here, we report that mice engineered to overexpress either the full-length murine B-Raf pseudogene Braf-rs1 or its pseudo "CDS" or "3' UTR" develop an aggressive malignancy resembling human diffuse large B cell lymphoma. We show that Braf-rs1 and its human ortholog, BRAFP1, elicit their oncogenic activity, at least in part, as competitive endogenous RNAs (ceRNAs) that elevate BRAF expression and MAPK activation in vitro and in vivo. Notably, we find that transcriptional or genomic aberrations of BRAFP1 occur frequently in multiple human cancers, including B cell lymphomas. Our engineered mouse models demonstrate the oncogenic potential of pseudogenes and indicate that ceRNA-mediated microRNA sequestration may contribute to the development of cancer. .
Copyright © 2015 Elsevier Inc. All rights reserved V体育安卓版. .
Figures





References
-
- Beard C, Hochedlinger K, Plath K, Wutz A, and Jaenisch R. (2006). Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–28. - PubMed
-
- Bier A, Oviedo-Landaverde I, Zhao J, Mamane Y, Kandouz M, and Batist G. (2009). Connexin43 pseudogene in breast cancer cells offers a novel therapeutic target. Mol. Cancer Ther 8, 786–793. - PubMed (V体育ios版)
V体育官网入口 - Publication types
- VSports手机版 - Actions
- "V体育官网" Actions
MeSH terms
- Actions (VSports注册入口)
- VSports - Actions
- Actions (VSports app下载)
- Actions (VSports)
Substances
- Actions (VSports)
Grants and funding
LinkOut - more resources
Full Text Sources
"VSports最新版本" Other Literature Sources
Molecular Biology Databases
"VSports在线直播" Research Materials
"V体育安卓版" Miscellaneous

