DNA barcoding reveals diverse growth kinetics of human breast tumour subclones in serially passaged xenografts
- PMID: 25532760
- PMCID: VSports在线直播 - PMC4284657
- DOI: 10.1038/ncomms6871
DNA barcoding reveals diverse growth kinetics of human breast tumour subclones in serially passaged xenografts (VSports在线直播)
Abstract
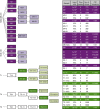
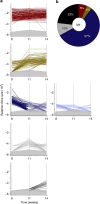
Genomic and phenotypic analyses indicate extensive intra- as well as intertumoral heterogeneity in primary human malignant cell populations despite their clonal origin. Cellular DNA barcoding offers a powerful and unbiased alternative to track the number and size of multiple subclones within a single human tumour xenograft and their response to continued in vivo passaging. Using this approach we find clone-initiating cell frequencies that vary from ~1/10 to ~1/10,000 cells transplanted for two human breast cancer cell lines and breast cancer xenografts derived from three different patients. For the cell lines, these frequencies are negatively affected in transplants of more than 20,000 cells. Serial transplants reveal five clonal growth patterns (unchanging, expanding, diminishing, fluctuating or of delayed onset), whose predominance is highly variable both between and within original samples. This study thus demonstrates the high growth potential and diverse growth properties of xenografted human breast cancer cells VSports手机版. .
VSports手机版 - Figures




References (V体育官网入口)
-
- Visvader J. E. & Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 28, 1143–1158 (2014). - PMC (VSports在线直播) - PubMed
-
- Meacham C. E. & Morrison S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013). - VSports注册入口 - PMC - PubMed
-
- Eirew P. et al.. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat. Med. 14, 1384–1389 (2008). - "V体育官网入口" PubMed
-
- Lim E. et al.. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 15, 907–913 (2009). - PubMed
Publication types
MeSH terms (V体育安卓版)
- "V体育ios版" Actions
- Actions (V体育2025版)
- "VSports" Actions
- "V体育2025版" Actions
Grants and funding
"V体育平台登录" LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

