Expanded identification and characterization of mammalian circular RNAs
- PMID: 25070500
- PMCID: PMC4165365 (V体育官网入口)
- DOI: 10.1186/s13059-014-0409-z (V体育ios版)
Expanded identification and characterization of mammalian circular RNAs
"VSports app下载" Abstract
Background: The recent reports of two circular RNAs (circRNAs) with strong potential to act as microRNA (miRNA) sponges suggest that circRNAs might play important roles in regulating gene expression. However, the global properties of circRNAs are not well understood. VSports手机版.
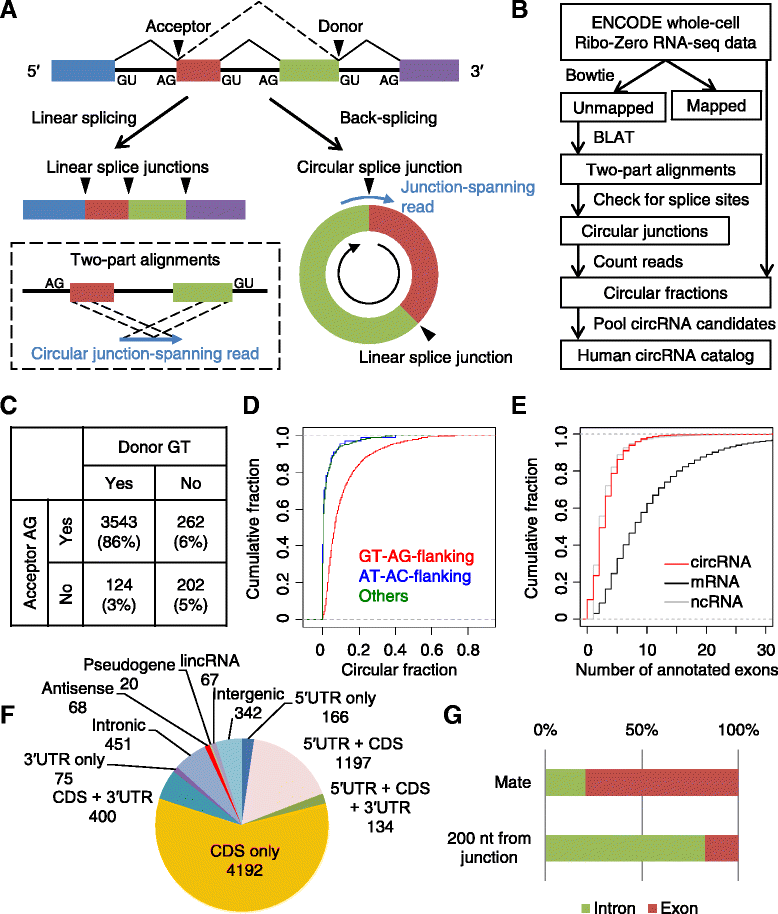
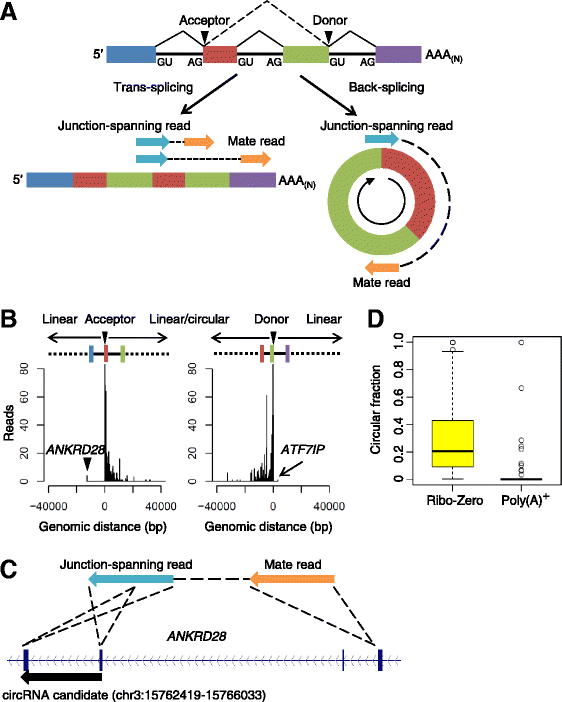
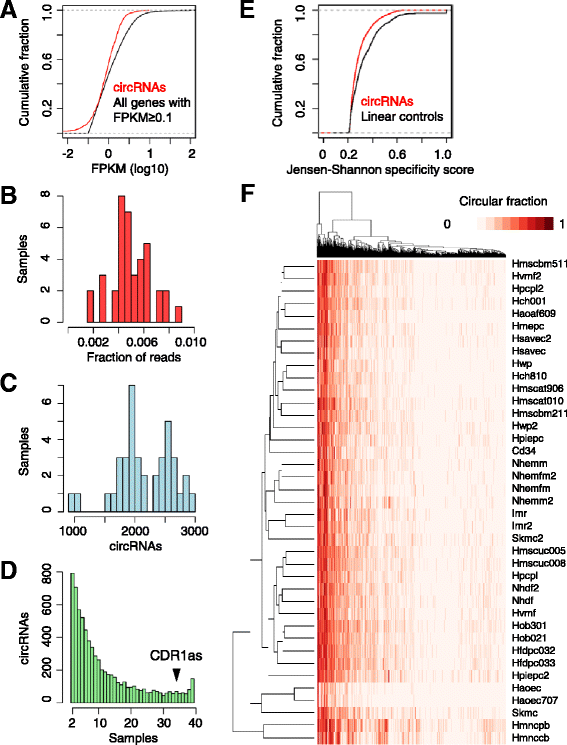
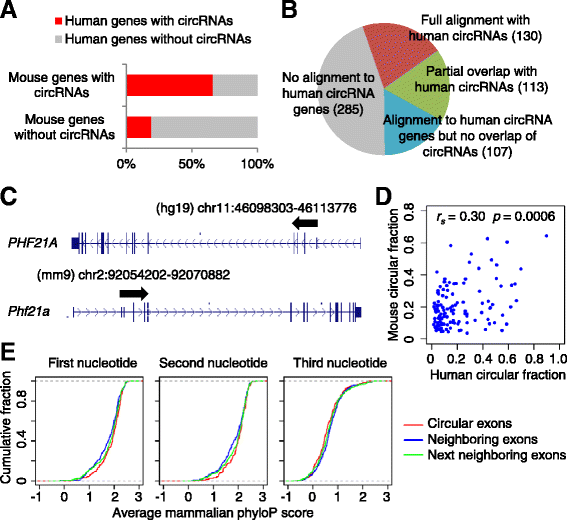
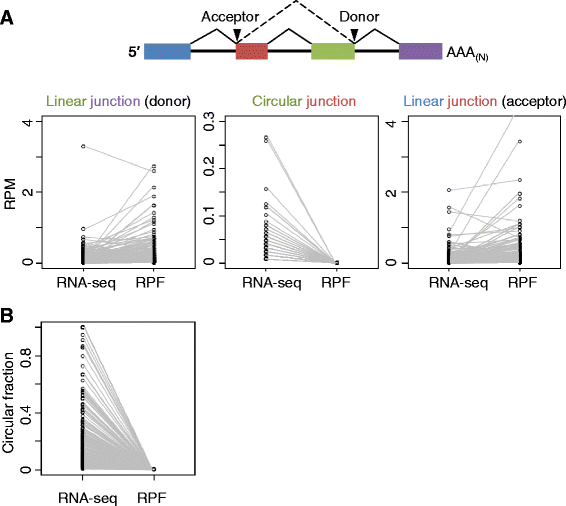
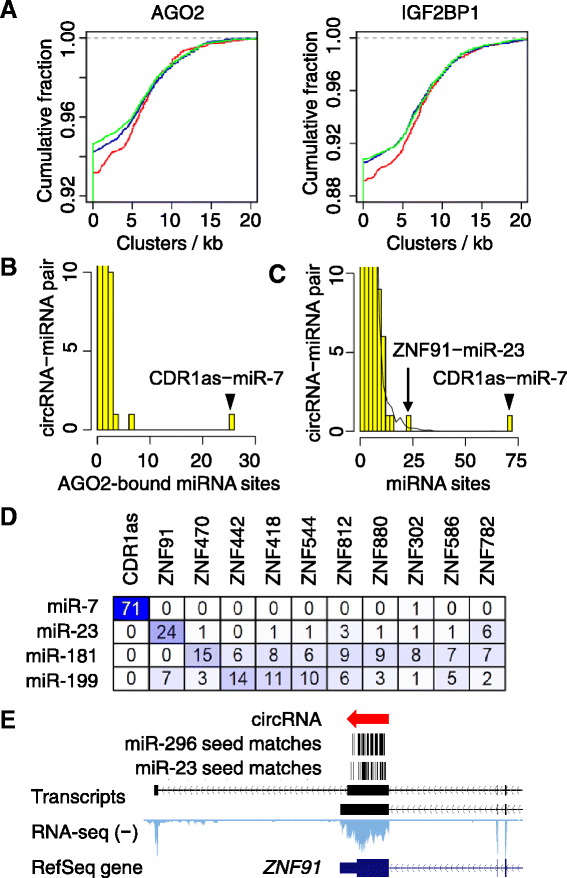
Results: We developed a computational pipeline to identify circRNAs and quantify their relative abundance from RNA-seq data. Applying this pipeline to a large set of non-poly(A)-selected RNA-seq data from the ENCODE project, we annotated 7,112 human circRNAs that were estimated to comprise at least 10% of the transcripts accumulating from their loci. Most circRNAs are expressed in only a few cell types and at low abundance, but they are no more cell-type-specific than are mRNAs with similar overall expression levels. Although most circRNAs overlap protein-coding sequences, ribosome profiling provides no evidence for their translation. We also annotated 635 mouse circRNAs, and although 20% of them are orthologous to human circRNAs, the sequence conservation of these circRNA orthologs is no higher than that of their neighboring linear exons. The previously proposed miR-7 sponge, CDR1as, is one of only two circRNAs with more miRNA sites than expected by chance, with the next best miRNA-sponge candidate deriving from a gene encoding a primate-specific zinc-finger protein, ZNF91. V体育安卓版.
Conclusions: Our results provide a new framework for future investigation of this intriguing topological isoform while raising doubts regarding a biological function of most circRNAs. V体育ios版.
Figures
References
-
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. - DOI - PMC - PubMed
-
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. - DOI - PMC - PubMed
Publication types
- Actions (V体育安卓版)
MeSH terms
- Actions (V体育官网)
- VSports最新版本 - Actions
- "V体育官网" Actions
- VSports最新版本 - Actions
- "VSports手机版" Actions
Substances
- VSports在线直播 - Actions
- "VSports最新版本" Actions
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

