Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition
- PMID: 25002527
- PMCID: PMC4135628
- DOI: 10.1128/MCB.00221-14
Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition
Erratum in
-
Correction for Ye et al., Nrf2- and ATF4-Dependent Upregulation of xCT Modulates the Sensitivity of T24 Bladder Carcinoma Cells to Proteasome Inhibition.Mol Cell Biol. 2015 Jul;35(13):2366. doi: 10.1128/MCB.00404-15. Mol Cell Biol. 2015. PMID: 26044038 Free PMC article. No abstract available.
"VSports在线直播" Abstract
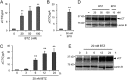
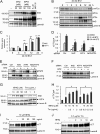
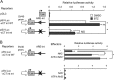
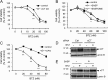
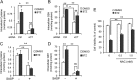
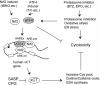
The ubiquitin-proteasome pathway degrades ubiquitinated proteins to remove damaged or misfolded protein and thus plays an important role in the maintenance of many important cellular processes. Because the pathway is also crucial for tumor cell growth and survival, proteasome inhibition by specific inhibitors exhibits potent antitumor effects in many cancer cells. xCT, a subunit of the cystine antiporter system xc (-), plays an important role in cellular cysteine and glutathione homeostasis. Several recent reports have revealed that xCT is involved in cancer cell survival; however, it was unknown whether xCT affects the cytotoxic effects of proteasome inhibitors. In this study, we found that two stress-inducible transcription factors, Nrf2 and ATF4, were upregulated by proteasome inhibition and cooperatively enhance human xCT gene expression upon proteasome inhibition. In addition, we demonstrated that the knockdown of xCT by small interfering RNA (siRNA) or pharmacological inhibition of xCT by sulfasalazine (SASP) or (S)-4-carboxyphenylglycine (CPG) significantly increased the sensitivity of T24 cells to proteasome inhibition. These results suggest that the simultaneous inhibition of both the proteasome and xCT could have therapeutic benefits in the treatment of bladder tumors VSports手机版. .
Copyright © 2014, American Society for Microbiology. All Rights Reserved. V体育安卓版.
Figures




References
-
- Voorhees PM, Dees EC, O'Neil B, Orlowski RZ. 2003. The proteasome as a target for cancer therapy. Clin. Cancer Res. 9:6316–6325 - PubMed
-
- Adams J. 2004. The development of proteasome inhibitors as anticancer drugs. Cancer Cell 5:417–421. 10.1016/S1535-6108(04)00120-5 - DOI (V体育ios版) - PubMed
Publication types
MeSH terms
- "VSports最新版本" Actions
- Actions (VSports手机版)
- V体育平台登录 - Actions
- V体育安卓版 - Actions
- "V体育ios版" Actions
- "V体育官网" Actions
- V体育2025版 - Actions
- VSports注册入口 - Actions
- Actions (V体育安卓版)
- "VSports" Actions
- V体育官网入口 - Actions
- Actions (V体育官网)
- Actions (VSports手机版)
Substances
- Actions (VSports注册入口)
- "VSports在线直播" Actions
- Actions (VSports)
- V体育ios版 - Actions
- V体育ios版 - Actions
- "V体育安卓版" Actions
- V体育官网入口 - Actions
- Actions (V体育官网入口)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
